Field Studies of Microbial Removal from Stormwater by Bioretention Cells with Fly-Ash Amendment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Sampling Methods
2.3. Laboratory Analysis
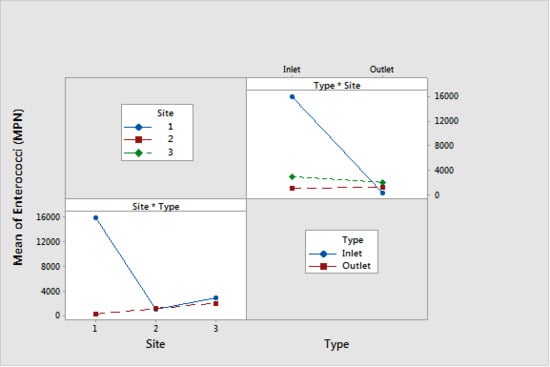
2.4. Statistical Analysis
3. Results
3.1. Basic Parameters
3.2. Microbial Concentrations and Removal
3.3. Comparison of Fly-Ash Amended Bioretention Cells in Grove, Oklahoma to Sand Cells in Current Literature
3.4. Conclusions and Recommendations for Future Work
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zhang, L.; Seagren, E.; Davis, A.; Karns, J. The capture and destruction of Escherichia coli from simulated urban runoff using conventional bioretention media and iron oxide-coated sand. Water Environ. Res. 2010, 82, 701–713. [Google Scholar] [CrossRef] [PubMed]
- Bradley, I.; Straub, A.; Maraccini, P.; Markazi, S.; Nguyen, T. Iron oxide amended biosand filters for virus removal. Water Res. 2011, 45, 4501–4510. [Google Scholar] [CrossRef] [PubMed]
- Schoonover, J.E.; Lockacy, B.G. Land cover impacts on stream nutrients and fecal coliform in the lower Piedmont of West Georgia. J. Hydrol. 2006, 331, 371–382. [Google Scholar] [CrossRef]
- Line, D.E.; White, N.M.; Kirby-Smith, W.W.; Potts, J.D. Fecal coliform export from four coastal North Carolina areas. J. Am. Water Resour. Assoc. 2008, 44, 606–617. [Google Scholar] [CrossRef]
- Lehner, P.; Aponte Clark, G.P.; Cameron, D.M.; Frank, A.G. Stormwater Strategies: Community Responses to Runoff Pollution. Nat. Resour. Def. Counc. 1999. Available online: http://www.nrdc.org/water/pollution/storm/stoinx.asp (accessed on 15 August 2015).
- Yakirevick, A.; Pachepsky, Y.; Guber, A.K.; Gish, T.J.; Shelton, D.R.; Cho, K.H. Modeling transport of Escherichia coli in a creek during and after artificial high flow events: Three year study and analysis. Water Resour. 2013, 47, 2676–2688. [Google Scholar] [CrossRef] [PubMed]
- Quilliam, R.; Jamieson, J.; Oliver, D. Seaweeds and plastic debris can influence the survival of faecal indicator organisms in beach environments. Mar. Pollut. Bull. 2014, 84, 201–207. [Google Scholar] [CrossRef] [PubMed]
- United States Environmental Protection Agency. Low Impact Development, 2000. Available online: http://www.lowimpactdevelopment.org/pubs/LID_litreview.pdf (accessed on 15 August 2015).
- United States Environmental Protection Agency. Water: Best Management Practices: Bioretention. Available online: http://water.epa.gov/polwaste/swbmp/Bioretention-Rain-Gardens.cfm (accessed on 25 May 2015).
- United States Environmental Protection Agency. Storm Water Technology Fact Sheet—Bioretention. EPA832-F-99–012, 1999. Available online: http://water.epa.gov/scitech/wastetech/upload/2002_06_28_mtb_biortn.pdf (accessed on 15 August 2015).
- Davis, A.P.; Shokouhian, M.; Sharma, H.; Minami, C. Laboratory Study of Biological Retention (Bioretention) for Urban Stormwater Management. Water Environ. Res. 2001, 73, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.P.; Shokouhian, M.; Sharma, H.; Minami, C. Water Quality Improvement through Bioretention Media: Nitrogen and Phosphorus Removal. Water Environ. Res. 2006, 78, 284–293. [Google Scholar] [CrossRef] [PubMed]
- Hunt, W.F.; Lord, W.G. Urban Waterways: Bioretention Performance, Design, Construction and Maintenance, 2006. Available online: http://www.bae.ncsu.edu/stormwater/PublicationFiles/Bioretention2006.pdf (accessed on 15 August 2015).
- Low Impact Development (LID) Center, INC. Bioretention—Pollutant Filtering. Available online: http://www.lid-stormwater.net/bio_benefits.htm (accessed on 15 August 2015).
- Davis, A.; Hunt, W.; Traver, R.; Clar, M. Bioretention Technology: Overview of Current Practice and Future Needs. J. Environ. Eng. 2009, 135, 109–117. [Google Scholar] [CrossRef]
- Roy-Poirier, A.; Champagne, P.; Filion, Y. Bioretention processes for phosphorus pollution control. Environ. Rev. 2010, 18, 159–173. [Google Scholar] [CrossRef]
- Jin, Y.; Chus, Y.; Li, Y. Virus removal and transport in saturated and unsaturated sand columns. J. Contam. Hydrol 2000, 43, 118–128. [Google Scholar] [CrossRef]
- Hathaway, J.M.; Hunt, W.E.; Wright, J.D.; Jadlocki, S.J. Field Evaluation of Indicator Bacteria Removal by Stormwater BMPs in North Carolina. World Environ. Water Resour. Congr. 2009, 1–10. [Google Scholar] [CrossRef]
- Park, S.J.; Lee, C.G.; Kim, S.B.; Chang, Y.Y.; Yang, J.K. Bacterial removal in flow-through columns packed with iron-manganese bimetallic oxide-coated sand. J. Environ. Sci. Health Part A Toxic/Hazard. Subst. Environ. Eng. 2012, 47, 1364–1371. [Google Scholar] [CrossRef] [PubMed]
- Potts, M. Desiccation tolerance of prokaryotes. Microbiol. Mol. Biol. Rev. 1994, 58, 755–805. [Google Scholar]
- Garbrecht, K.; Fox, G.A.; Guszman, J.A.; Alexander, D. E. coli Transport through Soil Columns: Implications for Bioretention Cell Removal Efficiency. Trans. ASABE 2009, 52, 481–486. [Google Scholar] [CrossRef]
- Hunt, W.F.; Smith, J.T.; Jadlocki, S.J.; Hathaway, J.M.; Eubanks, P.R. Pollutant removal and peak flow mitigation by a bioretention cell in urban Charlotte, NC. J. Environ. Eng. 2008, 134, 403–408. [Google Scholar] [CrossRef]
- Chavez, R.A.; Brown, G.O.; Coffman, R.R.; Storm, D.E. Design, construction and lessons learned from Oklahoma bioretention cell demonstration project. Appl. Eng. Agric. 2015, 31, 1–9. [Google Scholar]
- Brown, G.O.; Storm, D.E.; Chavez, R.A. Grand Lake, Oklahoma Watershed Implementation Project, Task 5.3 Bioretention Cell Design, Evaluation and Technology Demonstration; EPA Grant #C9–996100–12, Report to the Oklahoma Conservation Commission; Biosystems and Agricultural Engineering, Oklahoma State University: Stillwater, OK, USA, December 2008; p. 64, (and appendix folio). [Google Scholar]
- Zhang, W.; Brown, G.O.; Storm, D.E. Enhancement of heavy metals retention in sandy soil by amendment with fly ash. Trans. Am. Soc. Agric. Biol. Eng. 2008, 51, 1247–1254. [Google Scholar] [CrossRef]
- Zhang, W.; Brown, G.O.; Storm, D.E.; Zhang, H. Fly ash amended sand as filter media in bioretention cells to improve phosphorous removal. Water Environ. Res. 2008, 80, 507–516. [Google Scholar] [CrossRef] [PubMed]
- SevenMulti™. pH and EC Meter Operating Instructions; SevenMulti™: Toledo, OH, USA, 2012. [Google Scholar]
- ASTM D3977-97. Standard Test Methods for Determining Sediment Concentration in Water Samples, Method B, ASTM International: West Conshohocken, PA, USA, 2013.
- HACH. 2100Q and 2100Qis Basic User Manual, 2nd ed.; HACH: Loveland, CO, USA, 2013. [Google Scholar]
- IDEXX. IDEXX Enterolert Test Method for the Detection of Enterococci in Water; IDEXX: Westbrook, ME, USA, 2014. [Google Scholar]
- IDEXX. IDEXX Colilert Test Method for the Detection of Total Coliforms and E. coli in Water; IDEXX: Westbrook, ME, USA, 2013. [Google Scholar]
- United States Environmental Protection Agency. Recreational Water Quality Criteria; 820-F-12–058; Office of Water Quality: Washington, DC, USA, 2012.

| Site | Area (m2) | Volume (m3) | Drainage Area (Hectares) | Latitude and Longitude | Land Cover |
|---|---|---|---|---|---|
| Elm Creek Plaza (ECP) | 63 | 128 | 0.62 | 36.579643 −94.768417 | Paved |
| Grand Lake Association (GLA) | 172 | 435 | 0.76 | 36.610923 −94.8033817 | Paved/Turf |
| Grove High School (GHS) | 149 | 161 | 0.26 | 36.5779781 −94.7555676 | Paved |
| Composition | Content (%) |
|---|---|
| SiO2 | 38.1 |
| Al2O3 | 18.4 |
| Fe2O3 | 5.93 |
| MnO | 0.02 |
| MgO | 5.43 |
| CaO | 22.9 |
| Na2O | 1.82 |
| K2O | 0.56 |
| Ti2O | 1.39 |
| P2O5 | 1.37 |
| BaO | 0.69 |
| Cr2O3 | 0.01 |
| SrO | 0.30 |
| Loss on ignition | 0.69 |
| Other | 2.40 |
| Total | 100.0 |
| Site | Flume Characteristics | ||
|---|---|---|---|
| Inflow | Outflow | Overflow | |
| Elm Creek Plaza (ECP) | 0.3 m H flume | Palmer Bowlus flumes | Rectangular Concrete Weir |
| Grand Lake Association (GLA) | 0.46 m H flume | Palmer Bowlus flumes | Rectangular Concrete Weir |
| Grove High School (GHS) | 0.46 m H flume | Palmer Bowlus flumes | Rectangular Concrete Weir |
| Site | Elm Creek Plaza (ECP) | Grand Lake Association (GLA) | Grove High School (GHS) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | Standard Deviation | Range [High, Low] | Mean | Standard Deviation | Range [high, low] | Mean | Standard Deviation | Range [high, low] | |
| Flow Reduction (%) | 73 | 12 | [91, 47] | −1200 | 3329 | [80, −12,639] | 8 | 86 | [69, −220] |
| Storm Sampled (% Inlet) | 94 | 6 | [100, 91] | 110 | 34 | [172, 82] | 96 | 4 | [100, 84] |
| Storm Sampled (%Underdrain) | 84 | 21 | [100, 40] | 84 | 22 | [100, 54] | 85 | 16 | [98, 51] |
| pH (Inlet) | 6.8 | 0.8 | [3.7, 7.6] | 7.1 | 0.3 | [7.4, 6.2] | 6.8 | 0.7 | [8.5, 5.5] |
| pH (Underdrain) | 7.7 | 0.2 | [7.1, 7.9] | 7.9 | 0.2 | [8.3, 7.5] | 7.6 | 0.2 | [7.8, 7.3] |
| Electric Conductivity (EC) Inlet (µmhos/cm) | 74 | 26 | [159, 43] | 95 | 24 | [146, 67] | 160 | 238 | [805, 37] |
| Electric Conductivity (EC) Underdrain (µmhos/cm) | 210 | 37 | [305, 148] | 330 | 87 | [393, 61.6] | 175 | 29 | [240, 138] |
| Total Suspended Solids (TSS) Inlet (mg/L) | 117 | 73 | [251, 23] | 84 | 105 | [337, 12] | 78 | 74 | [258, 0] |
| Total Suspended Solids (TSS) Underdrain (mg/L) | 44 | 27 | [87, 0] | 27 | 28 | [80, 0] | 37 | 32 | [90, 0] |
| Turbidity Inlet (NTU) | 67 | 52 | [150, 0] | 9 | 4 | [15, 3] | 17 | 16 | [46, 0] |
| Turbidity Underdrain (NTU) | 7 | 5 | [14, 0] | 4 | 3 | [9, 1] | 3 | 2 | [5, 0] |
| Site | Elm Creek Plaza (ECP) | Grand Lake Association (GLA) | Grove High School (GHS) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | Standard Deviation | Range [High, Low] | Mean | Standard Deviation | Range [High, Low] | Mean | Standard Deviation | Range [High, Low] | |
| E. coli Inlet (MPN/100 mL) | 1600 | 1940 | [6900, 10] | 4900 | 7700 | [26,000, 104] | 1800 | 4700 | [18,000, <DL] |
| E. coli Underdrain (MPN/100 mL) | 810 | 1200 | [3700, <DL] | 310 | 380 | [1300, <DL] | 2000 | 3000 | [9200, 104] |
| Underdrain Met E. coli Recreation Limit (126/100 mL) | 5/23 | 5/14 | 5/16 | ||||||
| Enterococci Inlet (MPN/100 mL) | 3130 | 4200 | [20,000, 67] | 15,000 | 10,000 | [24,000, 52] | 3400 | 6300 | [1400, 40] |
| Enterococci Underdrain (MPN/100 mL) | 2100 | 3600 | [16,000, <DL] | 350 | 440 | [1300, <40] | 800 | 1700 | [5800, 20] |
| Underdrain Met Enterococci Recreation Limit (35/100 mL) | 1/23 | 1/14 | 2/16 | ||||||
| Coliphage Inlet (PFU/100 mL) | 14 | 22 | [67, <DL] | 7 | 11 | [33, <DL] | 5 | 10 | [17, 0] |
| Coliphage Underdrain (PFU/100 mL) | 9 | 23 | [100, <DL] | 2 | 5 | [17, <DL] | 4 | 10 | [<DL] |
| Site | Elm Creek Plaza (ECP) n = 20 | Grand Lake Association (GLA) n = 12 | Grove High School (GHS) n = 6 |
|---|---|---|---|
| E. coli Change in Concentration inlet to underdrain (%) | 51 | 94 | 22 |
| E. coli Mass Removal inlet to underdrain (%) | 91 | 39 | 58 |
| Did not meet E. coli limit on underdrain sample | 14/20 | 7/12 | 4/6 |
| Enterococci Change in Concentration inlet to underdrain (%) | 30 | 98 | −9 |
| Enterococci Mass Removal inlet to underdrain (%) | 81 | 95 | 20 |
| Did not meet Enterococci limit on underdrain sample | 19/20 | 11/12 | 5/6 |
| Coliphage Change in Concentration inlet to underdrain (%) | 25 | 75 | 100 |
| Coliphage Mass Removal inlet to underdrain (%) | 78 | 81 | 100 |
| Response Variable | Factor | p-Value | Mean | Tukey’s Multiple Comparison | |
|---|---|---|---|---|---|
| Enterococci (MPN) | Type | Inlet | <0.001 | 6700 | A |
| Underdrain | 1200 | B | |||
| Site | 1 | <0.001 | 8200 | A | |
| 3 | 2500 | B | |||
| 2 | 1200 | B | |||
| Media Type * Site Interaction | <0.001 | N/A | N/A | ||
| E. coli (MPN) | Type | Inlet | <0.001 | 3600 | N/A |
| Underdrain | 1400 | N/A | |||
| Site | 2 | 0.199 | 3400 | N/A | |
| 1 | 2800 | N/A | |||
| 3 | 1300 | N/A | |||
| Coliphage (PFU) | Type | Inlet | 0.495 | 7 | N/A |
| Underdrain | 4 | N/A | |||
| Site | 3 | 0.199 | 10 | N/A | |
| 1 | 4 | N/A | |||
| 2 | 1 | N/A | |||
| Pathogen | Paired t-Test | Mann-Whitney |
|---|---|---|
| p value * | p value * | |
| E. coli | 0.026 | 0.026 |
| Enterococci | 0.001 | <0.001 |
| Coliphage | 0.478 | 0.166 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Youngblood, S.; Vogel, J.; Brown, G.; Storm, D.; McLemore, A.; Kandel, S. Field Studies of Microbial Removal from Stormwater by Bioretention Cells with Fly-Ash Amendment. Water 2017, 9, 526. https://doi.org/10.3390/w9070526
Youngblood S, Vogel J, Brown G, Storm D, McLemore A, Kandel S. Field Studies of Microbial Removal from Stormwater by Bioretention Cells with Fly-Ash Amendment. Water. 2017; 9(7):526. https://doi.org/10.3390/w9070526
Chicago/Turabian StyleYoungblood, Sheila, Jason Vogel, Glenn Brown, Daniel Storm, Alex McLemore, and Saroj Kandel. 2017. "Field Studies of Microbial Removal from Stormwater by Bioretention Cells with Fly-Ash Amendment" Water 9, no. 7: 526. https://doi.org/10.3390/w9070526
APA StyleYoungblood, S., Vogel, J., Brown, G., Storm, D., McLemore, A., & Kandel, S. (2017). Field Studies of Microbial Removal from Stormwater by Bioretention Cells with Fly-Ash Amendment. Water, 9(7), 526. https://doi.org/10.3390/w9070526