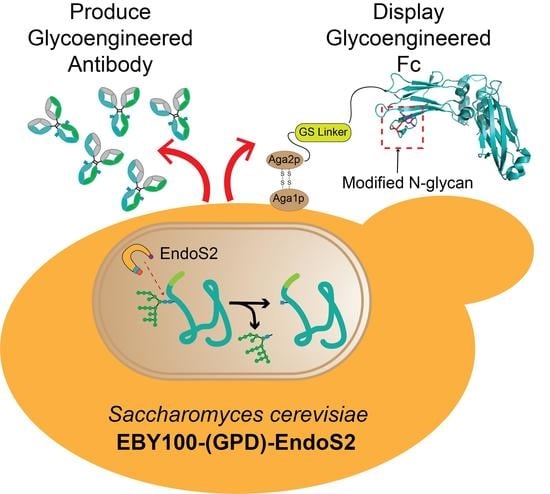
Expression and Display of Glycoengineered Antibodies and Antibody Fragments with an Engineered Yeast Strain
Abstract
:1. Introduction
2. Materials and Methods
2.1. Material
2.2. Strain and Media
2.3. Cloning of Fc into the Yeast Surface Display Plasmid pYD1
2.4. Creation and Validation of the EBY100-EndoS2 Strains
2.5. Protein Sample Preparation
2.6. Flow Cytometry to Determine Fc Surface Expression
2.7. Expression and Purification of IgG1 Fc and Rituximab
2.8. Purification Using Concanavalin A Column
2.9. Cloning the Heavy Chain and Light Chain of Rituximab into pESC-TRP1
2.10. Mass Spectrometry
2.11. Surface Plasmon Resonance
2.12. Flow Cytometry for Fab Functionality with Raji B-Cells
3. Results
3.1. IgG1 Fc with a Hydrolyzed N-Glycan Expressed on Yeast Binds GFP-FcγRIIIa
3.2. IgG1 Fc with a Truncated N-Glycan Is Secreted by EBY100-(GPD) EndoS2
3.3. Glycoengineered Rituximab Expressed with EBY100-(GPD) EndoS2
3.4. Purification of Full-Length Rituximab from the EBY100-(GPD) EndoS2 Strain
3.5. Functionality of Rituximab Isolated from EBY100-(GPD) EndoS2
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Duehr, J.; Wohlbold, T.J.; Oestereich, L.; Chromikova, V.; Amanat, F.; Rajendran, M.; Gomez-Medina, S.; Mena, I.; Tenoever, B.R.; García-Sastre, A.; et al. Novel Cross-Reactive Monoclonal Antibodies against Ebolavirus Glycoproteins Show Protection in a Murine Challenge Model vaccines and antiviral agents Crossm. J. Virol. 2017, 91, 652–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, P.; Chi, X.; Liu, G.; Zhang, G.; Chen, Z.; Liu, Y.; Fang, T.; Li, J.; Banadyga, L.; He, S.; et al. Potent neutralizing monoclonal antibodies against Ebola virus isolated from vaccinated donors. mAbs 2020, 12, 1742457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef] [PubMed]
- Gaebler, C.; Wang, Z.; Lorenzi, J.C.C.; Muecksch, F.; Finkin, S.; Tokuyama, M.; Cho, A.; Jankovic, M.; Schaefer-Babajew, D.; Oliveira, T.Y.; et al. Evolution of antibody immunity to SARS-CoV-2. Nature 2021, 591, 639–644. [Google Scholar] [CrossRef]
- Tay, M.Z.; Wiehe, K.; Pollara, J. Antibody-Dependent Cellular Phagocytosis in Antiviral Immune Responses. Front. Immunol. 2019, 10, 332. [Google Scholar] [CrossRef]
- van Erp, E.A.; Luytjes, W.; Ferwerda, G.; van Kasteren, P.B. Fc-Mediated Antibody Effector Functions during Respiratory Syncytial Virus Infection and Disease. Front. Immunol. 2019, 10, 548. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.; Brown, D.; Reed, C.; Chung, S.; Lutman, J.; Stefanich, E.; Wong, A.; Stephan, J.-P.; Bayer, R. Production, characterization and pharmacokinetic properties of antibodies with N-linked Mannose-5 glycans. mAbs 2012, 4, 475–487. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Liu, Y.D.; Flynn, G.C. The effect of Fc glycan forms on human IgG2 antibody clearance in humans. Glycobiology 2008, 19, 240–249. [Google Scholar] [CrossRef]
- Liu, L.; Stadheim, A.; Hamuro, L.; Pittman, T.; Wang, W.; Zha, D.; Hochman, J.; Prueksaritanont, T. Pharmacokinetics of IgG1 monoclonal antibodies produced in humanized Pichia pastoris with specific glycoforms: A comparative study with CHO produced materials. Biologicals 2011, 39, 205–210. [Google Scholar] [CrossRef]
- Boune, S.; Hu, P.; Epstein, A.L.; Khawli, L.A. Principles of N-Linked Glycosylation Variations of IgG-Based Therapeutics: Pharmacokinetic and Functional Considerations. Antibodies 2020, 9, 22. [Google Scholar] [CrossRef]
- Shukla, A.A.; Wolfe, L.S.; Mostafa, S.S.; Norman, C. Evolving trends in mAb production processes. Bioeng. Transl. Med. 2017, 2, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Goetze, A.M.; Liu, Y.D.; Zhang, Z.; Shah, B.; Lee, E.; Bondarenko, P.V.; Flynn, G.C. High-mannose glycans on the Fc region of therapeutic IgG antibodies increase serum clearance in humans. Glycobiology 2011, 21, 949–959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moremen, K.W.; Tiemeyer, M.; Nairn, A. Vertebrate protein glycosylation: Diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 2012, 13, 448–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalziel, M.; Crispin, M.; Scanlan, C.N.; Zitzmann, N.; Dwek, R.A. Emerging Principles for the Therapeutic Exploitation of Glycosylation. Science 2014, 343, 1235681. [Google Scholar] [CrossRef]
- Vervecken, W.; Kaigorodov, V.; Callewaert, N.; Geysens, S.; De Vusser, K.; Contreras, R. In Vivo Synthesis of Mammalian-Like, Hybrid-Type N-Glycans in Pichia pastoris. Appl. Environ. Microbiol. 2004, 70, 2639–2646. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, S.R.; Bobrowicz, P.; Bobrowicz, B.; Davidson, R.C.; Li, H.; Mitchell, T.; Nett, J.H.; Rausch, S.; Stadheim, T.A.; Wischnewski, H.; et al. Production of Complex Human Glycoproteins in Yeast. Science 2003, 301, 1244–1246. [Google Scholar] [CrossRef]
- Callewaert, N.; Laroy, W.; Cadirgi, H.; Geysens, S.; Saelens, X.; Min Jou, W.; Contreras, R. Use of HDEL-Tagged Trichoderma Reesei Mannosyl Oligosaccharide 1, 2-α-D-Mannosidase for N-Glycan Engineering in Pichia Pastoris. FEBS Lett. 2001, 503, 173–178. [Google Scholar] [CrossRef]
- Chiba, Y.; Suzuki, M.; Yoshida, S.; Yoshida, A.; Ikenaga, H.; Takeuchi, M.; Jigami, Y.; Ichishima, E. Production of Human Compatible High Mannose-type (Man5GlcNAc2) Sugar Chains in Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 26298–26304. [Google Scholar] [CrossRef] [Green Version]
- Choi, B.-K.; Bobrowicz, P.; Davidson, R.C.; Hamilton, S.R.; Kung, D.H.; Li, H.; Miele, R.G.; Nett, J.H.; Wildt, S.; Gerngross, T.U. Use of combinatorial genetic libraries to humanize N-linked glycosylation in the yeast Pichia pastoris. Proc. Natl. Acad. Sci. USA 2003, 100, 5022–5027. [Google Scholar] [CrossRef] [Green Version]
- Bobrowicz, P.; Davidson, R.C.; Li, H.; Potgieter, T.I.; Nett, J.H.; Hamilton, S.R.; Stadheim, T.A.; Miele, R.G.; Bobrowicz, B.; Mitchell, T.; et al. Engineering of an artificial glycosylation pathway blocked in core oligosaccharide assembly in the yeast Pichia pastoris: Production of complex humanized glycoproteins with terminal galactose. Glycobiology 2004, 14, 757–766. [Google Scholar] [CrossRef] [Green Version]
- Nasab, F.P.; Aebi, M.; Bernhard, G.; Frey, A.D. A Combined System for Engineering Glycosylation Efficiency and Glycan Structure in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2013, 79, 997–1007. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Wang, X.; Yu, X.; Fu, L.; Liu, Y.; Ma, L.; Zhai, C. High-Level Expression of Endo-β-N-Acetylglucosaminidase H from Streptomyces plicatus in Pichia pastoris and Its Application for the Deglycosylation of Glycoproteins. PLoS ONE 2015, 10, e0120458. [Google Scholar] [CrossRef] [Green Version]
- Wildt, S.; Gerngross, T.U. The humanization of N-glycosylation pathways in yeast. Nat. Rev. Genet. 2005, 3, 119–128. [Google Scholar] [CrossRef]
- Mamedov, T.; Cicek, K.; Gulec, B.; Ungor, R.; Hasanova, G. In vivo production of non-glycosylated recombinant proteins in Nicotiana benthamiana plants by co-expression with Endo-β-N-acetylglucosaminidase H (Endo H) of Streptomyces plicatus. PLoS ONE 2017, 12, e0183589. [Google Scholar] [CrossRef] [Green Version]
- Bennett, L.D.; Yang, Q.; Berquist, B.R.; Giddens, J.P.; Ren, Z.; Kommineni, V.; Murray, R.P.; White, E.L.; Holtz, B.R.; Wang, L.-X.; et al. Implementation of Glycan Remodeling to Plant-Made Therapeutic Antibodies. Int. J. Mol. Sci. 2018, 19, 421. [Google Scholar] [CrossRef] [Green Version]
- Kao, D.; Danzer, H.; Collin, M.; Groß, A.; Eichler, J.; Stambuk, J.; Lauc, G.; Lux, A.; Nimmerjahn, F. A Monosaccharide Residue Is Sufficient to Maintain Mouse and Human IgG Subclass Activity and Directs IgG Effector Functions to Cellular Fc Receptors. Cell Rep. 2015, 13, 2376–2385. [Google Scholar] [CrossRef] [Green Version]
- Subedi, G.P.; Barb, A.W. The Structural Role of Antibody N-Glycosylation in Receptor Interactions. Structure 2015, 23, 1573–1583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collin, M. EndoS, a novel secreted protein from Streptococcus pyogenes with endoglycosidase activity on human IgG. EMBO J. 2001, 20, 3046–3055. [Google Scholar] [CrossRef] [PubMed]
- Sjögren, J.; Struwe, W.; Cosgrave, E.; Rudd, P.M.; Stervander, M.; Allhorn, M.; Hollands, A.; Nizet, V.; Collin, M. EndoS2 is a unique and conserved enzyme of serotype M49 group A Streptococcus that hydrolyses N-linked glycans on IgG and α1-acid glycoprotein. Biochem. J. 2013, 455, 107–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sjogren, J.; Cosgrave, E.F.J.; Allhorn, M.; Nordgren, M.; Björk, S.; Olsson, F.; Fredriksson, S.; Collin, M. EndoS and EndoS2 hydrolyze Fc-glycans on therapeutic antibodies with different glycoform selectivity and can be used for rapid quantification of high-mannose glycans. Glycobiology 2015, 25, 1053–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes, A.M.V.; Carmo, T.S.; Carvalho, L.S.; Bahia, F.M.; Parachin, N.S. Comparison of Yeasts as Hosts for Recombinant Protein Production. Microorganisms 2018, 6, 38. [Google Scholar] [CrossRef] [Green Version]
- Lin-Cereghino, J.; Wong, W.W.; Xiong, S.; Giang, W.; Luong, L.T.; Vu, J.; Johnson, S.D.; Lin-Cereghino, G.P. Condensed protocol for competent cell preparation and transformation of the methylotrophic yeast Pichia pastoris. BioTechniques 2005, 38, 44–48. [Google Scholar] [CrossRef] [Green Version]
- Boder, E.T.; Wittrup, K.D. Yeast Surface Display for Screening Con1binatorial Polypeptide Libraries. Nat. Biotechnol. 1997, 15, 553–557. [Google Scholar] [CrossRef]
- Subedi, G.P.; Johnson, R.W.; Moniz, H.A.; Moremen, K.W.; Barb, A. High Yield Expression of Recombinant Human Proteins with the Transient Transfection of HEK293 Cells in Suspension. J. Vis. Exp. 2015, e53568. [Google Scholar] [CrossRef] [Green Version]
- Subedi, G.P.; Hanson, Q.M.; Barb, A.W. Restricted Motion of the Conserved Immunoglobulin G1 N-Glycan Is Essential for Efficient FcγRIIIa Binding. Structure 2014, 22, 1478–1488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mei, M.; Zhai, C.; Li, X.; Zhou, Y.; Peng, W.; Ma, L.; Wang, Q.; Iverson, B.L.; Zhang, G.; Yi, L. Characterization of aromatic residue–controlled protein retention in the endoplasmic reticulum of Saccharomyces cerevisiae. J. Biol. Chem. 2017, 292, 20707–20719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voth, W.P.; Richards, J.D.; Shaw, J.M.; Stillman, D.J. Yeast Vectors for Integration at the HO Locus. Nucleic Acids Res. 2001, 29, e59. [Google Scholar] [CrossRef] [Green Version]
- Gnügge, R.; Liphardt, T.; Rudolf, F. A shuttle vector series for precise genetic engineering ofSaccharomyces cerevisiae. Yeast 2015, 33, 83–98. [Google Scholar] [CrossRef]
- Uchański, T.; Zögg, T.; Yin, J.; Yuan, D.; Wohlkönig, A.; Fischer, B.; Rosenbaum, D.M.; Kobilka, B.K.; Pardon, E.; Steyaert, J. An improved yeast surface display platform for the screening of nanobody immune libraries. Sci. Rep. 2019, 9, 382. [Google Scholar] [CrossRef]
- Wang, S.; Cho, Y.K. Yeast surface display of full-length human microtubule-associated protein tau. Biotechnol. Prog. 2019, 36, e2920. [Google Scholar] [CrossRef] [PubMed]
- Rakestraw, J.A.; Sazinsky, S.L.; Piatesi, A.; Antipov, E.; Wittrup, K.D. Directed evolution of a secretory leader for the improved expression of heterologous proteins and full-length antibodies inSaccharomyces cerevisiae. Biotechnol. Bioeng. 2009, 103, 1192–1201. [Google Scholar] [CrossRef] [Green Version]
- Fishman, J.B.; Berg, E.A. Protein A and Protein G Purification of Antibodies. Cold Spring Harb. Protoc. 2019, 2019. [Google Scholar] [CrossRef]
- Deisenhofer, J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-A resolution. Biochemistry 1981, 20, 2361–2370. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-P.; Tsai, T.-I.; Cheng, T.; Shivatare, V.S.; Wu, C.-Y.; Wong, C.-H. Glycoengineering of antibody (Herceptin) through yeast expression and in vitro enzymatic glycosylation. Proc. Natl. Acad. Sci. USA 2018, 115, 720–725. [Google Scholar] [CrossRef] [Green Version]
- Li, A.; Liu, Z.; Li, Q.; Yu, L.; Wang, D.; Deng, X. Construction and characterization of bidirectional expression vectors inSaccharomyces cerevisiae. FEMS Yeast Res. 2008, 8, 6–9. [Google Scholar] [CrossRef] [Green Version]
- Rougé, L.; Chiang, N.; Steffek, M.; Kugel, C.; Croll, T.I.; Tam, C.; Estevez, A.; Arthur, C.P.; Koth, C.M.; Ciferri, C.; et al. Structural biology. Science 2020, 367, 1224–1230. [Google Scholar] [CrossRef]
- Shields, R.L.; Lai, J.; Keck, R.; O’Connell, L.Y.; Hong, K.; Meng, Y.G.; Weikert, S.H.A.; Presta, L.G. Lack of Fucose on Human IgG1 N-Linked Oligosaccharide Improves Binding to Human FcγRIII and Antibody-dependent Cellular Toxicity. J. Biol. Chem. 2002, 277, 26733–26740. [Google Scholar] [CrossRef] [Green Version]
- Subedi, G.P.; Barb, A.W. The immunoglobulin G1 N-glycan composition affects binding to each low affinity Fc γ receptor. mAbs 2016, 8, 1512–1524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allhorn, M.; Olin, A.I.; Nimmerjahn, F.; Collin, M. Human IgG/FcγR Interactions Are Modulated by Streptococcal IgG Glycan Hydrolysis. PLoS ONE 2008, 3, e1413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Zhang, H.; Liu, X.; Wang, P.G.; Qi, Q. Influence of N-Glycosylation on Saccharomyces cerevisiae Morphology: A Golgi Glycosylation Mutant Shows Cell Division Defects. Curr. Microbiol. 2007, 55, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Horwitz, A.H.; Chang, C.P.; Better, M.; Hellstromt, K.E.; Robinson, R.R. Secretion of functional antibody and Fab fragment from yeast cells. Proc. Natl. Acad. Sci. USA 1988, 85, 8678–8682. [Google Scholar] [CrossRef] [Green Version]
- Kelley, B. Industrialization of mAb production technology: The bioprocessing industry at a crossroads. mAbs 2009, 1, 443–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Tong, X.; Yang, Q.; Giddens, J.; Wang, L.-X. Glycosynthase Mutants of Endoglycosidase S2 Show Potent Transglycosylation Activity and Remarkably Relaxed Substrate Specificity for Antibody Glycosylation Remodeling. J. Biol. Chem. 2016, 291, 16508–16518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, W.; Giddens, J.; Fan, S.-Q.; Toonstra, C.; Wang, L.-X. Chemoenzymatic Glycoengineering of Intact IgG Antibodies for Gain of Functions. J. Am. Chem. Soc. 2012, 134, 12308–12318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parsons, T.B.; Struwe, W.B.; Gault, J.; Yamamoto, K.; Taylor, T.A.; Raj, R.; Wals, K.; Mohammed, S.; Robinson, C.V.; Benesch, J.L.P.; et al. Optimal Synthetic Glycosylation of a Therapeutic Antibody. Angew. Chem. 2016, 128, 2407–2413. [Google Scholar] [CrossRef] [PubMed] [Green Version]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shenoy, A.; Yalamanchili, S.; Davis, A.R.; Barb, A.W. Expression and Display of Glycoengineered Antibodies and Antibody Fragments with an Engineered Yeast Strain. Antibodies 2021, 10, 38. https://doi.org/10.3390/antib10040038
Shenoy A, Yalamanchili S, Davis AR, Barb AW. Expression and Display of Glycoengineered Antibodies and Antibody Fragments with an Engineered Yeast Strain. Antibodies. 2021; 10(4):38. https://doi.org/10.3390/antib10040038
Chicago/Turabian StyleShenoy, Anjali, Srisaimaneesh Yalamanchili, Alexander R. Davis, and Adam W. Barb. 2021. "Expression and Display of Glycoengineered Antibodies and Antibody Fragments with an Engineered Yeast Strain" Antibodies 10, no. 4: 38. https://doi.org/10.3390/antib10040038
APA StyleShenoy, A., Yalamanchili, S., Davis, A. R., & Barb, A. W. (2021). Expression and Display of Glycoengineered Antibodies and Antibody Fragments with an Engineered Yeast Strain. Antibodies, 10(4), 38. https://doi.org/10.3390/antib10040038