Chiral Heterocycle-Based Receptors for Enantioselective Recognition
Abstract
:1. Introduction
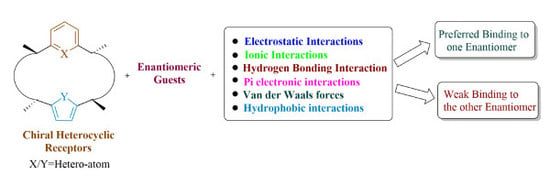
2. Components of Chiral Molecular Recognition
2.1. Different Types of Forces
2.2. Non-Covalent Interactions
2.2.1. Electrostatic Interactions
Hydrogen Bonding Interaction
2.2.2. π-Electronic Interactions
π-π Interaction
Cation–π Interaction
2.2.3. Van der Waals Forces
2.2.4. Hydrophobic Interactions
3. Analytical Tool for Chiral Recognition
3.1. Fluorescence
- F0 = Initial emission intensity of the host
- F = Intensity of the host after addition of analyte (Guest)
- Ksv = Stern-Volmer constant/Stability constant for the complex
- [Q] = Concentration of the analyte (Guest)
- I0 = Initial emission intensity of the host
- I = Intensity of the host after addition of analyte (Guest)
- K = Stability constant for the complex
- a, b = Constants terms
- [M] = Concentration of the analyte (Guest)
3.2. UV-Vis Absorption
- [H]0 and [G]0 are total concentrations of host and guest, respectively,
- Δε is the change of molar extinction coefficient between the free and complexed host,
- ΔA represents the absorption change of host upon the addition of opposite guest enantiomers.
3.3. Circular Dichroism (CD)
3.4. Nuclear Magnetic Resonance (NMR) Analysis
3.5. High Performance Liquid Chromatography (HPLC)
3.6. Mass Spectrometry
- (1)
- IRIS > 1.0 means that the given chiral host binds more strongly (R)-enantiomer of the guest, hence indicating the (R)-enantiomer preference; the larger IR/IS−dn value corresponds to the higher degree of chiral recognition of the host.
- (2)
- In contrast, IRIS < 1.0 means that the given chiral host binds more strongly (S)-enantiomer of the guest, indicating the (S)-enantiomer preference with the opposite tendency for the IR/IS−dn value.
- (3)
- IRIS = 1.0 ± 0.05 means that the given chiral host cannot differentiate the chirality of the guest.
3.7. Electrochemical Methods
Cyclic Voltammetry
4. Special Features of Heterocyclic Receptors for Enantioselective Recognition
- Heterocycles have unshared electron pairs present on the heteroatoms useful for the three points hydrogen bond formation, especially when chiral ammonium cations are studied, with the chiral guest molecule.
- Heterocycles possess a permanent dipole responsible for the charge-dipole electrostatic interactions.
- The aromatic heterocycles have π electrons to facilitate the corresponding π-π stacking interaction and cation-π binding with a chiral aromatic guest molecule.
- The conformational rigidity is increased by the presence of heterocyclic ring, which imparts a good deal of preorganization of the chiral host suiting a guest molecule.
- The aliphatic heterocyclic ring system may assist the hydrophobic interaction with a chiral guest molecule.
- The heterocyclic ring may additionally influence the steric interaction responsible for chiral discrimination.
5. Chiral Hosts with Six Member Heterocycle/s
5.1. Nitrogen Containing Six Member Heterocycle/s
Pyridine Ring
5.2. Oxygen Containing Six Member Heterocycle
6. Five Member Heterocycles Containing Receptors
6.1. Imidazole Ring Containing Receptors
6.2. Benzimidazole Ring Containing Receptors
6.3. Triazole Ring Containing Receptors
6.4. Benzo-Fused Furan Heterocycles Containing Receptors
6.5. Receptors with Nitrogen and Oxygen Containing Five Membered Heterocycles
7. Miscellaneous Heterocyclic Receptors
8. Summary
Acknowledgments
Conflicts of Interest
References
- Zupancic, V.; Kikelj, D. Chiral drugs in pharmaceutical industry-development and applications. Farmacevtski Vestnik 2001, 52, 259–272. [Google Scholar]
- Gao, Y.; Boschetti, E.; Guerrier, L. Synthesis and separation of optically active compounds. Part I. Ann. Pharm. Fr. 1994, 52, 184–203. [Google Scholar] [PubMed]
- Hembury, G.A.; Borovkov, V.V.; Inoue, Y. Chirality sensing supramolecular systems. Chem. Rev. 2008, 108, 1–73. [Google Scholar] [CrossRef] [PubMed]
- Ball, P. Giving life a hand. Chem. World 2007, 4, 30–31. [Google Scholar]
- Talele, T.T. Natural-products-inspired use of the gem-dimethyl group in medicinal chemistry. J. Med. Chem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Blaskovich, M.A.T. Unusual amino acids in medicinal chemistry. J. Med. Chem. 2016, 59, 10807–10836. [Google Scholar] [CrossRef] [PubMed]
- Karnik, A.V.; Patil, S.T.; Patnekar, S.S.; Semwal, A. A convenient route to optically active γ-substituted γ-lactones. New J. Chem. 2004, 28, 1420–1422. [Google Scholar] [CrossRef]
- Karnik, A.V.; Kamath, S.S. Cascade enantioselective synthesis of γ-aryl-γ-butyrolactones with a delayed stereoselective step. Tetrahedron 2008, 64, 2992–2996. [Google Scholar] [CrossRef]
- Tawde, T.S.; Wagh, S.J.; Sapre, J.V.; Khose, V.N.; Badani, P.M.; Karnik, A.V. Reversal of enantioselectivity induced by the achiral part of an organocatalyst in a Diels-Alder reaction. Tetrahedron Asymmetry 2016, 27, 130–135. [Google Scholar] [CrossRef]
- Buchbauer, G.; Shafii-Tabatabai, A. Enones of (+)- and (−)-3-pinanone: Influence of chirality on flavour. Flavour Fragr. J. 2003, 18, 441–445. [Google Scholar] [CrossRef]
- Leffingwell, J.C.; Leffingwell, D. Chiral chemistry in flavours & fragrances. Spec. Chem. Mag. 2011, 31, 30–33. [Google Scholar]
- Pietropaolo, A. Ideas in Chemistry and Molecular Sciences: Where Chemistry Meets Life; Pignataro, B., Ed.; Wiley: Hoboken, NJ, USA, 2010; pp. 293–312. [Google Scholar]
- Venkatakrishnarao, D.; Sahoo, C.; Mamonov, E.A.; Novikov, V.B.; Mitetelo, N.V.; Naraharisetty, S.G.; Murzina, T.V.; Chandrasekar, R. Chiral organic photonics: Self-assembled micro-resonators for an enhanced circular dichroism effect in the non-linear optical signal. J. Mater. Chem. C 2017, 5, 12349–12353. [Google Scholar] [CrossRef]
- Cram, D.J. The design of molecular hosts, guests, and their complexes. Science 1988, 240, 760–767. [Google Scholar] [CrossRef] [PubMed]
- Bao, W.; Wang, Z.; Li, Y. Synthesis of chiral ionic liquids from natural amino acids. J. Org. Chem. 2003, 68, 591–593. [Google Scholar] [CrossRef] [PubMed]
- Bako, P.; Fenichel, L.; Toke, L. A Novel diaza-crown ether and cryptands from glucopyranose and their complex-forming properties. Eur. J. Org. Chem. 1990, 1990, 1161–1164. [Google Scholar] [CrossRef]
- Bako, P.; Vizvardi, K.; Toppet, S.; Eycken, E.; Hoornaert, G.; Fenichel, L.; Toke, L. Synthesis, extraction ability and application in asymmetric synthesis of aza crown ethers derived from d-glucose. Tetrahedron 1998, 54, 14975–14988. [Google Scholar] [CrossRef]
- Bradshaw, J.S.; Huszthy, P.; McDaniel, C.W.; Oue, M.; Zhu, C.Y.; Izatt, R.M.; Lifson, S. Enantiomeric recognition of organic ammonium salts by chiral pyridino-18-crown-6 ligands: A short review. J. Coord. Chem. 1992, 27, 105–114. [Google Scholar] [CrossRef]
- Fernandez-Gonzalez, A.; Guardia, L.; Badía-Laíño, R.; Díaz-García, M. Mimicking molecular receptors for antibiotics–analytical implications. Trends Anal. Chem. 2006, 25, 949–957. [Google Scholar] [CrossRef]
- Kaushansky, K. Small molecule mimics of hematopoietic growth factors: Improving on Mother Nature? Leukemia 2001, 15, 673–674. [Google Scholar] [CrossRef] [PubMed]
- Dickert, F.L. Biomimetic receptors and sensors. Sensors 2014, 14, 22525–22531. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, K.; Jung, J.; Shin, S.; Ahn, K. Unprecedented Chiral Molecular Recognition in a C3-Symmetric Environment. J. Am. Chem. Soc. 2002, 124, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Seo, J.; Yim, E.; Jin, Y.; Song, S.; Suh, H. Enantiomeric recognition in host-guest complexation using chiral bis-pyridino-18-crown-6 ethers, by electrospray ionization mass spectrometry (ESI-MS) enantiomer-labelled (EL) guest method. Bull. Korean Chem. Soc. 2008, 9, 1069–1072. [Google Scholar]
- Lu, Q.; Hou, J.; Wang, J.; Xu, B.; Zhang, J.; Yu, X. Multichannel Chromogenic and Chiral Anions Recognition by Imidazolium Functionalized BINOL Derivatives. Chin. J. Chem. 2013, 31, 641–650. [Google Scholar] [CrossRef]
- Hasan, M.; Khose, V.N.; Pandey, A.D.; Borovkov, V.; Karnik, A.V. Tailor-Made Supramolecular Chirogenic System Based on Cs-Symmetric Rigid Organophosphoric Acid Host and Amino Alcohols: Mechanistic Studies, Bulkiness Effect, and Chirality Sensing. Org. Lett. 2016, 18, 440–443. [Google Scholar] [CrossRef] [PubMed]
- Artz, S.P.; de Grandpre, M.P.; Cram, D.J. Host-guest complexation. Search for new chiral hosts. J. Org. Chem. 1985, 50, 1486–1496. [Google Scholar] [CrossRef]
- Ozer, H.; Kocakaya, S.O.; Akgun, A.; Hosgoren, H.; Togrul, M. The enantiomeric recognition of chiral organic ammonium salts by chiral pyridino-macrocycles bearing aminoalcohol subunits. Tetrahedron Asymmetry 2009, 20, 1541–1546. [Google Scholar] [CrossRef]
- Shahgaldian, P.; Pieles, U.; Hegner, M. Enantioselective recognition of phenylalanine by a chiral amphiphilic macrocycle at the air-water interface: A copper-mediated mechanism. Langmuir 2005, 21, 6503–6507. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Dong, L.; Zhang, J.; Li, J.; Jiang, L.; Huang, Y.; Qin, S.; Hu, C.; Yu, X. Imidazolium-functionalized binol as a multifunctional receptor for chromogenic and chiral anion recognition. Org. Lett. 2009, 11, 669–672. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Hua, W. Synthesis and characterization of pyridine-based polyamido-polyester optically active macrocycles and enantiomeric recognition for d- and l-amino acid methyl ester hydrochloride. J. Org. Chem. 2000, 65, 2933–2938. [Google Scholar] [CrossRef] [PubMed]
- Granda, J.M.; Jurczak, J. Sweet anion receptors: Recognition of chiral carboxylate anions by d-glucuronic-acid-decorated diindolylmethane. Org. Lett. 2013, 15, 4730–4733. [Google Scholar] [CrossRef] [PubMed]
- De la Torre, M.F.; Campos, E.G.; Gonzalez, S.; Moran, J.R.; Caballero, M.C. Binding properties of an abiotic receptor for complexing carboxylates of α-heterocyclic and α-keto acids. Tetrahedron 2001, 57, 3945–3950. [Google Scholar] [CrossRef]
- Tejeda, A.; Oliva, A.I.; Simon, L.; Grande, M.; Caballero, M.; Moran, J.R. A macrocyclic receptor for the chiral recognition of hydroxycarboxylates. Tetrahedron Lett. 2000, 41, 4563–4566. [Google Scholar] [CrossRef]
- You, J.; Yu, X.; Zhang, G.; Xiang, Q.; Lan, J.; Xie, R. Novel chiral imidazole cyclophane receptors: Synthesis and enantioselective recognition for amino acid derivatives. Chem. Commun. 2001, 0, 1816–1817. [Google Scholar] [CrossRef]
- Pandey, A.; Mohammed, H.; Karnik, A. Chiral benzimidazole-derived mono azacrowns: Synthesis and enantiomer recognition studies with chiral amines and their ammonium salts. Tetrahedron Asymmetry 2013, 24, 706–712. [Google Scholar] [CrossRef]
- Pandey, A.D.; Mohammed, H.; Pissurlenkar, R.S.; Karnik, A.V. Size-induced chiral discrimination switching by (S)-(−)-2-(α-hydroxyethyl)-benzimidazole-derived azacrowns. ChemPlusChem 2015, 80, 475–479. [Google Scholar] [CrossRef]
- Upadhyay, S.P.; Pissurlenkar, R.S.; Coutinho, E.C.; Karnik, A.V. Furo-fused BINOL based crown as a fluorescent chiral sensor for enantioselective recognition of phenylethylamine and ethyl ester of Valine. J. Org. Chem. 2007, 72, 5709–5714. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Guo, H.; James, T.D.; Zhao, J. Enantioselective recognition of mandelic acid by a 3,6-dithiophen-2-yl-9H-carbazole-based chiral fluorescent bisboronic acid sensor. J. Org. Chem. 2011, 76, 5685–5695. [Google Scholar] [CrossRef] [PubMed]
- Shcherbakova, E.G.; Brega, V.; Minami, T.; Sheykhi, S.; James, T.D.; Anzenbacher, P. Toward fluorescence-based high-throughput screening for enantiomeric excess in amines and amino acid derivatives. Chem. Eur. J. 2016, 22, 10074–10080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jo, H.H.; Lin, C.Y.; Anslyn, E.V. Rapid optical methods for enantiomeric excess analysis: From enantioselective indicator displacement assays to exciton-coupled circular dichroism. Acc. Chem. Res. 2014, 47, 2212–2221. [Google Scholar] [CrossRef] [PubMed]
- Velazquez, H.A.; Hamelberg, D. Electrochemical detection. In Chemosensors: Principles, Strategies, and Applications, 1st ed.; Wang, B., Anslyn, E.V., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2011. [Google Scholar]
- Lynnf, B.C.; Tsesarskaja, M.; Schall, O.F.; Hernandez, J.C.; Watanabe, S.; Takahashit, T.; Kaifer, A.; Gokel, G.W. Hydrogen bonding in macrocyclic receptor systems. Supramol. Chem. 1993, 1, 253–260. [Google Scholar] [CrossRef]
- Muehldorf, A.V.; Engen, D.V.; Warner, J.C.; Hamilton, A.D. Aromatic-aromatic interactions in molecular recognition: A family of artificial receptors for thymine that shows both face-to-face and edge-to-face orientations. J. Am. Chem. Soc. 1988, 110, 6561–6562. [Google Scholar] [CrossRef]
- Mecozzi, S.; West, A.P.; Dougherty, D.A. Cation-π interactions in aromatics of biological and medicinal interest: Electrostatic potential surfaces as a useful qualitative guide. Proc. Natl. Acad. Sci. USA 1996, 93, 10566–10571. [Google Scholar] [CrossRef] [PubMed]
- Price, S.L.; Stone, A.J. The electrostatic interactions in van der Waals complexes involving aromatic molecules. J. Chem. Phys. 1987, 86, 2859–2868. [Google Scholar] [CrossRef]
- Gibb, B.C. Van der waals interactions and the hydrophobic effect. In Chemosensors: Principles, Strategies, and Applications, 1st ed.; Wang, B., Anslyn, E.V., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2011. [Google Scholar]
- Mei, X.; Wolf, C. Enantioselective sensing of chiral carboxylic acids. J. Am. Chem. Soc. 2004, 126, 14736–14737. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.; Khose, V.N.; Mori, T.; Borovkov, V.; Karnik, A.V. Sui Generis Helicene-Based Supramolecular Chirogenic System: Enantioselective Sensing, Solvent Control, and Application in Chiral Group Transfer Reaction. ACS Omega 2017, 2, 592–598. [Google Scholar] [CrossRef]
- Li, G.; Cao, J.; Zong, W.; Lei, X.; Tan, R. Enantiodiscrimination of carboxylic acids using the diphenylprolinol NMR chiral solvating agents. Org. Chem. Front. 2016, 3, 96–102. [Google Scholar] [CrossRef]
- Fukui, F.; Fukushi, Y. NMR Determinations of the Absolute Configuration of α-Chiral Primary Amines. Org. Lett. 2010, 12, 2856–2859. [Google Scholar] [CrossRef] [PubMed]
- Parker, D. NMR determination of enantiomeric purity. Chem. Rev. 1991, 91, 1441–1457. [Google Scholar] [CrossRef]
- Davankov, V.A. Separation of enantiomeric compounds using chiral HPLC systems. A brief review of general principles, advances, and development trends. Chromatographia 1989, 27, 475–482. [Google Scholar] [CrossRef]
- Kim, J.; Lee, J.; Lee, S.; Seo, J.; Hong, J.; Suh, H. Chiral molecular recognition in fast atom bombardment mass spectrometry (FAB-MS) enantiomerlabeled (EL) guest method using new chiral bis-pyridino-18-crown-6. Bull. Korean Chem. Soc. 2002, 23, 543–544. [Google Scholar]
- Bayly, S.R.; Chen, G.Z.; Beer, P.D. Electrochemical Detection. In Chemosensors: Principles, Strategies, and Applications, 1st ed.; Wang, B., Anslyn, E.V., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2011. [Google Scholar]
- Shaw, S.A.; Aleman, P.; Vedejs, E. Development of chiral nucleophilic pyridine catalysts: Applications in asymmetric quaternary carbonsynthesis. J. Am. Chem. Soc. 2003, 125, 13368–13369. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Fujioka, S.; Sekiguchi, Y.; Kotsuki, H. A new class of chiral pyrrolidine-pyridine conjugate base catalysts for use in asymmetric Michael addition reactions. J. Am. Chem. Soc. 2004, 126, 9558–9559. [Google Scholar] [CrossRef] [PubMed]
- Ager, D.J.; Prakash, I.; Schaad, D.R. 1,2-amino alcohols and their heterocyclic derivatives as chiral auxiliaries in asymmetric synthesis. Chem. Rev. 1996, 96, 835–876. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.C. Asymmetric Catalysis with “Planar-Chiral” Derivatives of 4-(Dimethylamino)pyridine. Acc. Chem. Res. 2004, 37, 542–547. [Google Scholar] [CrossRef] [PubMed]
- Jolly, S.T.; Bradshaw, J.S.; Izatt, R.M. Synthetic chiral macrocyclic crown ligands: A short review. J. Heterocycl. Chem. 1982, 19, 3–18. [Google Scholar] [CrossRef]
- Bradshaw, J.S.; Peter Huszthy, P.; Redd, J.T.; Zhang, X.X.; Wang, T.; Hathaway, J.K.; Young, J.; Izatt, R.M. Enantiomeric recognition of chiral ammonium salts by chiral pyridino- and pyrimidino-18-crown-6 ligands: Effect of structure and solvents. Pure Appl. Chem. 1995, 67, 691–695. [Google Scholar] [CrossRef]
- Chu, I.; Dearden, D.V.; Bradshaw, J.S.; Huszthy, P.; Izatt, R.M. Chiral host-guest recognition in an ion-molecule reaction. J. Am. Chem. Soc. 1993, 115, 4318–4320. [Google Scholar] [CrossRef]
- Hellier, P.C.; Bradshaw, J.S.; Young, J.J.; Zhang, X.X.; Izatt, R.M. Chiral pyridine-based macrobicyclic clefts: Synthesis and enantiomeric recognition of ammonium salts. J. Org. Chem. 1996, 61, 7270–7275. [Google Scholar] [CrossRef] [PubMed]
- Asakawa, M.; Brown, C.L.; Pasini, D.; Stoddart, J.F.; Wyatt, P.G. Enantioselective recognition of amino acids by axially-chiral π-electron-deficient receptors. J. Org. Chem. 1996, 61, 7234–7235. [Google Scholar] [CrossRef] [PubMed]
- Somogyi, L.; Huszthy, P.; Bradshaw, J.S.; Izatt, R.M.; Hollosi, M. Enantiomeric recognition of aralkyl ammonium salts by chiral pyridino-18-crown-6 ligands: Use of circular dichroism spectroscopy. Chirality 1997, 9, 545–549. [Google Scholar] [CrossRef]
- Gavin, J.A.; Garcia, M.E.; Benesi, A.J.; Mallouk, T.E. Chiral molecular recognition in a tripeptide benzylviologen cyclophane host. J. Org. Chem. 1998, 63, 7663–7669. [Google Scholar] [CrossRef]
- Du, C.; You, J.; Yu, X.; Liu, C.; Lan, J.; Xie, R. Homochiral molecular tweezers as hosts for the highly enantioselective recognition of amino acid derivatives. Tetrahedron Asymmetry 2003, 14, 3651–3656. [Google Scholar] [CrossRef]
- Chen, X.; Du, D.; Hua, W. Synthesis of novel chiral polyamide macrocycles containing pyridyl side-arms and their molecular recognition properties. Tetrahedron Asymmetry 2003, 14, 999–1007. [Google Scholar] [CrossRef]
- Ema, T.; Tanida, D.; Sakai, T. Versatile and practical macrocyclic reagent with multiple hydrogen-bonding sites for chiral discrimination in NMR. J. Am. Chem. Soc. 2007, 129, 10591–10596. [Google Scholar] [CrossRef] [PubMed]
- Heckel, T.; Winkel, A.; Wilhelm, R. Chiral ionic liquids based on nicotine for the chiral recognition of carboxylic acids. Tetrahedron Asymmetry 2013, 24, 1127–1133. [Google Scholar] [CrossRef]
- Ma, F.; Ai, L.; Shen, X.; Zhang, C. New macrocyclic compound as chiral shift reagent for carboxylic acids. Org. Lett. 2007, 9, 125–127. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Huang, Z.; Chen, S.; Li, K.; Yu, X.; Pu, L. Enantioselective gel collapsing: A new means of visual chiral sensing. J. Am. Chem. Soc. 2010, 132, 7297–7299. [Google Scholar] [CrossRef] [PubMed]
- Seker, S.; Barıs, D.; Arslan, N.; Turgut, Y.; Lu, N.P.; Togrul, M. Synthesis of rigid and C2-symmetric pyridino-15-crown-5 type macrocycles bearing diamide-diester functions: Enantiomeric recognition for chiral primary organoammonium perchlorate salts. Tetrahedron Asymmetry 2014, 25, 411–417. [Google Scholar] [CrossRef]
- Deniz, P.; Turgut, Y.; Togrul, M.; Hosgoren, H. Pyridine containing chiral macrocycles: Synthesis and their enantiomeric recognition for amino acid derivatives. Tetrahedron 2011, 67, 6227–6232. [Google Scholar] [CrossRef]
- Ghosh, K.; Majumdar, A. L-Amino acid derived pyridinium-based chiral compounds and their efficacy in chiral recognition of lactate. RSC Adv. 2015, 5, 24499–24506. [Google Scholar] [CrossRef]
- Khanvilkar, A.N.; Bedekar, A.V. Synthesis and characterization of chiral aza-macrocycles and study of their enantiomer recognition ability for organo-phosphoric acid and phosphonic acid derivatives by 31P NMR and fluorescence spectroscopy. Org. Biomol. Chem. 2016, 14, 2742–2748. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, S.; Pelaez, R.; Sanz, F.; Jimenez, M.B.; Moran, J.R.; Caballero, M.C. Macrocyclic chiral receptors toward enantioselective recognition of naproxen. Org. Lett. 2006, 8, 4679–4682. [Google Scholar] [CrossRef] [PubMed]
- Barnhill, D.K.; Sargent, A.L.; Allen, W.E. Participation of host spacer atoms in carboxylic acid binding: Implications for amino acid recognition. Tetrahedron 2005, 61, 8366–8371. [Google Scholar] [CrossRef]
- Su, X.; Luo, K.; Xiang, Q.; Lan, J.; Xie, R. Enantioselective recognitions of chiral molecular tweezers containing imidazoliums for amino acids. Chirality 2009, 21, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Luo, K.; Jiang, H.; You, J.; Xiang, Q.; Guoa, S.; Lan, J.; Xie, R. Chiral di-imidazolium molecular tweezers: Synthesis and enantioselective recognition for amino acid derivatives. Lett. Org. Chem. 2006, 3, 363–367. [Google Scholar]
- Munusamy, S.; Muralidharan, V.P.; Iyer, S.K. Enantioselective recognition of unmodified amino acids by ligand-displacement assays with in situ generated 1:1 Cu(II)-BINOL imidazole complex. Sens. Actuators B Chem. 2017, 250, 244–249. [Google Scholar] [CrossRef]
- Ghosh, K.; Sarkar, T. l-Valine derived benzimidazole based bis-urea in enantioselective fluorescence sensing of L-tartrate. Tetrahedron Lett. 2013, 54, 4568–4573. [Google Scholar] [CrossRef]
- Zhao, S.; Ito, S.; Ohba, Y.; Katagiri, H. Determination of the absolute configuration and identity of chiral carboxylic acids using a Cu(II) complex of pyridine-benzimidazole-based ligand. Tetrahedron Lett. 2014, 55, 2097–2100. [Google Scholar] [CrossRef]
- Sato, H.; Shizuma, M. Triazole-linked host compounds for chiral-discrimination toward amino acid ester guests. J. Oleo Sci. 2008, 57, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Miao, F.; Zhou, J.; Tian, D.; Li, H. Enantioselective recognition of mandelic acid with (R)-1,1-bi-2-naphthol-linked calix[4]arene via fluorescence and dynamic light scattering. Org. Lett. 2012, 14, 3572–3575. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Lu, J.; Liu, J.; Zheng, C.; Gao, Y.; Hu, J.; Ju, Y. A deoxycholic acid-based macrocycle: Recognition of mercury ion and cascade enantioselective sensing toward amino acids. Sens. Actuators B 2017, 241, 931–937. [Google Scholar] [CrossRef]
- Karnik, A.V.; Upadhyay, S.P.; Gangrade, M.G. [9,9′]Bi[naphtho(2,1-b)furanyl]-8,8′-diol, a furo-fused BINOL derivative: Synthesis, resolution and determination of absolute configuration. Tetrahedron Asymmetry 2006, 17, 1275–1280. [Google Scholar] [CrossRef]
- Upadhyay, S.P.; Karnik, A.V. Enantioselective synthesis of (R) and (S)-[9,9′]bi[naphtho(2,1-b)furanyl]-8,8′-diol. A furo-fused BINOL derivative. Tetrahedron Lett. 2007, 48, 317–318. [Google Scholar] [CrossRef]
- Kotwal, S.B.; Pandey, A.D.; Khose, V.N.; Karnik, A.V. A convenient route to enantiomerically enriched furo-fused BINOL derivative. Indian J. Chem. B 2015, 54, 940–943. [Google Scholar]
- Hasan, M.; Pandey, A.D.; Khose, V.N.; Mirgane, N.A.; Karnik, A.V. Sterically congested chiral 7,8-dioxa[6]helicene and its dihydro analogues: Synthesis, regioselective functionalization, and unexpected domino prins reaction. Eur. J. Org. Chem. 2015, 17, 3702–3712. [Google Scholar] [CrossRef]
- Ollevier, T. Iron bis(oxazoline) complexes in asymmetric catalysis. Catal. Sci. Technol. 2016, 6, 41–48. [Google Scholar] [CrossRef]
- Wolf, C.; Xu, H. Asymmetric catalysis with chiral oxazolidine ligands. Chem. Commun. 2011, 47, 3339–3350. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, K.; Kim, Y.K.; Shin, S.K.; Ahn, K.H. Crucial role of three-center hydrogen bonding in a challenging chiral molecular recognition. J. Am. Chem. Soc. 2003, 125, 13819–13824. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, S.; Seong, H.R.; Ahn, K.H. Breaking the C3-symmetry of chiral tripodal oxazolines: Enantio-discrimination of chiral organoammonium ions. J. Org. Chem. 2005, 70, 7227–7231. [Google Scholar] [CrossRef] [PubMed]
- Sambasivan, S.; Kim, S.; Choi, U.M.; Rhee, Y.M.; Ahn, K.H. C3-symmetric cage-like receptors: Chiral discrimination of R-chiral amines in a confined space. Org. Lett. 2010, 12, 4228–4231. [Google Scholar] [CrossRef] [PubMed]
- Oliva, A.I.; Simon, L.; Hernandez, J.V.; Muniz, F.M.; Lithgow, A.; Jimenez, A.; Moran, J.R. Enantioselective recognition of α-amino acid derivatives with a cis-tetrahydrobenzoxanthene receptor. J. Chem. Soc. Perkin Trans. 2002, 2, 1050–1052. [Google Scholar] [CrossRef]
- Roussel, C.; Roman, M.; Andreoli, F.; Del Rio, A.L.; Faure, R.; Vanthuyne, N. Non-racemic atropisomeric (thio)ureas as neutral enantioselective anion receptors for amino-acid derivatives: Origin of smaller Kass with thiourea than urea derivatives. Chirality 2006, 18, 762–771. [Google Scholar] [CrossRef] [PubMed]
- Satishkumar, S.; Periasamy, M. Chiral recognition of carboxylic acids by Troeger’s base derivatives. Tetrahedron Asymmetry 2009, 20, 2257–2262. [Google Scholar] [CrossRef]
- Schnopp, M.; Haberhauer, G. Highly selective recognition of α-chiral primary organoammonium ions by C3-symmetric peptide receptors. Eur. J. Org. Chem. 2009, 4458–4467. [Google Scholar] [CrossRef]
- Yu, S.; DeBerardinis, A.M.; Turlington, M.; Pu, L. Study of the Fluorescent Properties of partially hydrogenated 1,1′-Bi-2-naphthol-amine molecules and their use for enantioselective fluorescent recognition. J. Org. Chem. 2011, 76, 2814–2819. [Google Scholar] [CrossRef] [PubMed]
- Lichosyt, D.; Wasiłek, S.; Jurczak, J. Stereoselective chirality extension of syn,anti- and syn,syn-oxazine and stereochemical analysis of chiral 1,3-oxazines: Stereoselective total syntheses of (+)-1-deoxygalactonojirimycin and (−)-1-deoxygulonojirimycin. J. Org. Chem. 2016, 81, 7342–7348. [Google Scholar] [CrossRef] [PubMed]
- Konishi, A.; Nakaoka, K.; Maruyama, H.; Nakajima, H.; Eguchi, T.; Baba, A.; Yasuda, M. C3-symmetric boron lewis acid with a cage-shape for chiral molecular recognition and asymmetric catalysis. Chem. Eur. J. 2017, 23, 1273–1277. [Google Scholar] [CrossRef] [PubMed]


















































































| Entry | Substrate | Solvent | K2 (M−1) | Ka(L)/Ka(D) | −ΔG0 (kcal/mol) | ΔΔG0 (kcal/mol) b | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| (R)-14 | (R,R)-16 | (R)-14 | (R,R)-16 | (R)-14 | (R,R)-16 | (R)-14 | (R,R)-16 | |||
| 1 | L-Trp | H2O c | 2470 | ND d | 4.63 | |||||
| 2 | D-Trp | H2O c | 5860 | ND d | 0.42 | 5.14 | −0.51 | |||
| 3 | L-Trp OMe.HCl | H2O c | 753 | ND d | 3.92 | |||||
| 4 | D-Trp OMe.HCl | H2O c | 803 | ND d | 0.94 | 3.96 | −0.04 | |||
| 5 | N-Ac-L-Trp | A e | 20700 | 4280 | 5.89 | 4.95 | ||||
| 6 | N-Ac-D-Trp | A e | 2670 | 1080 | 7.75 | 3.96 | 4.67 | 4.14 | 1.22 | 0.81 |
| 7 | N-Ac-L-Tyr | B e | 10060 | 2340 | 5.45 | |||||
| 8 | N-Ac-D-Tyr | B e | 2125 | 1047 | 4.73 | 2.23 | 4.53 | 4.12 | 0.92 | 0.48 |
| 9 | N-Ac-L-Phe | A e | 1220 | 219 | 4.21 | 3.19 | ||||
| 10 | N-Ac-D-Phe | A e | 2260 | 137 | 0.54 | 1.60 | 4.57 | 2.91 | −0.36 | 0.28 |
| Guest | Ka (M−1) | Solvent System | Structure |
|---|---|---|---|
| (R)-DOPA (S)-DOPA | 39 ± 6 3 ± 1 | 17D2O:1acetone-d6:1DCl |  |
| (R)-Tryptophan (S)-Tryptophan | 5 ± 1 6 ± 1 | 17D2O:1acetone-d6:1DCl |  |
| Pindalol | 6 ± 1 | 17D2O:1acetone-d6:1DCl |  |
| Nandolol | 23 ± 3 | 10acetone-d6:3D2O |  |
| (R)-(−)-α-Methoxy Phenylacetic acid (S)-(+)-α-Methoxy Phenylacetic acid | 5 ± 1 8 ± 6 | 1acetone-d6:1D2O |  |
| N-(2-naphthyl)alaninate | 10 ± 1 | 9acetone-d6:4D2O |  |
| (S)-6-methoxy-α-methyl-2-naphthalene acetic acid | 9 ± 1 | 29D2O:11acetone-d6:1MeOD |  |
| Ligand | Amino Acid Methyl Ester Hydrochlorides, Cation | λmax (nm) | Δλ (nm) | I/I0 (Rel. Intensity) |
|---|---|---|---|---|
| 29a | 350 | |||
| d-Ala | 342 | 8 | 0.68 | |
| l-Ala | 350 | 0.68 | ||
| d-Phe | 350 | 50 | 0.84 | |
| l-Phe | 400 | 1.25 | ||
| d-His | 350 | 0 | 0.67 | |
| l-His | 350 | 0.66 | ||
| 29b | 392 | |||
| d-Ala | 362 | 12 | 1.08 | |
| l-Ala | 374 | 0.84 | ||
| d-Phe | 360 | 10 | 1.32 | |
| l-Phe | 350 | 1.21 | ||
| d-His | 362 | 38 | 0.91 | |
| l-His | 400 | 0.98 | ||
| 29c | 348 | |||
| d-Ala | 400 | 52 | 0.37 | |
| l-Ala | 348 | 0.85 | ||
| d-Phe | 352 | 48 | 0.69 | |
| l-Phe | 400 | 1.50 | ||
| d-His | 352 | 1 | 0.72 | |
| l-His | 351 | 0.77 | ||
| 29d | 392 | |||
| d-Ala | 354 | 12 | 1.19 | |
| l-Ala | 366 | 0.96 | ||
| d-Phe | 380 | 14 | 1.01 | |
| l-Phe | 366 | 1.19 | ||
| d-His | 350 | 16 | 1.96 | |
| l-His | 364 | 0.82 | ||
| 29e | 350 | |||
| d-Ala | 400 | 50 | 0.78 | |
| l-Ala | 450 | 4.69 | ||
| d-Phe | 350 | 50 | 0.84 | |
| l-Phe | 400 | 1.25 | ||
| d-His | 348 | 8 | 1.05 | |
| l-His | 340 | 0.99 |
| Host/Guest | 44 | 45 | 46 | 47 | 48 | 49 | 50 | 51 | 52 | 53 |
|---|---|---|---|---|---|---|---|---|---|---|
| 42 | 7.66 | 1.65 | 3.41 | 2.36 | 5.59 | 1.30 | 1.94 | 4.17 | 1.80 | 3.44 |
| 43 | 0.22 | 0.82 | 2.27 | 0.52 | 1.06 | 2.31 | 0.81 | 0.52 | 1.68 | 2.23 |
| Entry | Host | Guest b | Ka (dm3 mol−1) | KL/KD | −ΔG0 (kJ mol−1) | ΔΔG0 (kJ mol−1) c |
|---|---|---|---|---|---|---|
| 1 | 54 | l-Phe-OMe | 46.3 | 0.75 | 9.50 | 0.73 |
| 2 | 54 | d-Phe-OMe | 62.0 | 10.23 | ||
| 3 | 54 | l-Trp-OMe | 58.0 | 0.67 | 10.06 | 0.98 |
| 4 | 54 | d-Trp-OMe | 86.1 | 11.04 | ||
| 5 | 55 | l-Ala-OMe | 74.1 | 1.21 | 10.67 | −0.48 |
| 6 | 55 | d-Ala-OMe | 61.2 | 10.19 | ||
| 7 | 55 | l-Leu-OMe | 68.0 | 2.70 | 10.45 | −2.46 |
| 8 | 55 | d-Leu-OMe | 25.2 | 7.99 | ||
| 9 | 55 | l-Phe-OMe | 213 | 3.92 | 13.28 | −3.38 |
| 10 | 55 | d-Phe-OMe | 54.4 | 9.90 | ||
| 11 | 55 | l-Trp-OMe | 1280 | 7.90 | 17.72 | −5.12 |
| 12 | 55 | d-Trp-OMe | 162 | 12.60 | ||
| 13 | 56 | l-Leu-OMe | 59.9 | 2.0 | 10.14 | −1.71 |
| 14 | 56 | d-Leu-OMe | 30.0 | 8.43 | ||
| 15 | 57 | l-Leu-OMe | 36.8 | 1.33 | 8.93 | −0.72 |
| 16 | 57 | d-Leu-OMe | 27.7 | 8.23 |
| Entry | Salts | CDCl3 | CD3OD | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 19Fδ Mosher’s Acid | Δδ | 1H δ Mosher’s Acid | Δ δ | 1H δ Mandelic Acid | Δ δ | 19Fδ Mosher’s Acid | Δ δ | 1HδMosher’s Acid | Δ δ | ||
| 1 | - | −71.0 | - | 3.56q | - | 5.25 | - | −73.17 | - | 3.57q | - |
| 2 |  | −70.61/−70.63 | 5.9 | 3.51/3.50 | 7.5 | 4.99/5.01 | 5.7 | - | - | - | - |
| 3 |  | −70.86/−70.89 | 9.0 | 3.44/3.40 | 20.0 | 4.96/4.98 | 10.0 | −71.26/−71.29 | 9.1 | 3.55q/3.56q | 4.0 |
| 4 |  | −71.09 | 0 | 3.54/3.53 | 6.5 | 5.19 | 0 | −71.11/−71.13 | 6.6 | 3.56q/3.57q | 5.0 |
| 5 |  | −70.90/−70.92 | 7.2 | 3.49/3.47 | 10.0 | 4.95/4.96 | 5.4 | −71.20 | 0 | 3.56q | 0 |
| 6 |  | −71.02/−71.04 | 5.0 | 3.47/3.46 | 5.5 | 5.01/5.03 | 6.8 | −71.18 | 0 | 3.57q | 0 |
| 7 |  | −69.90/−70.57 | 7.5 | 3.45/3.42 | 17.0 | 5.20 | 0 | −71.64/−71.65 | 4.3 | 3.55q/3.56q | 2.0 |
| 8 |  | −70.67/−70.68 | 4.5 | 3.54/3.53 | 4.5 | 4.99/5.00 | 5.0 | −70.85/−70.88 | 8.1 | 3.58q/3.57q | 4.5 |
| 9 |  | −71.10 | 0 | 3.50/3.48 | 8.0 | 5.24/5.26 | 7.8 | −71.69/−71.70 | 5.5 | 3.57q/3.56q | 3.0 |
| Entry | Solvent | ΔΔ δ | (ppm) |
|---|---|---|---|
| 4 | 13 | ||
| 1 | CDCl3 | 0.62 | 0.39 |
| 2 | CDCl3/C6D6 (10%) | 0.62 | 0.39 |
| 3 | CDCl3/Acetone-d6 (10%) | 0.53 | 0.27 |
| 4 | CDCl3/CD3OD (10%) | 0.43 | 0.21 |
| 5 | CDCl3/DMSO-d6 (10%) | 0.13 | 0.04 |
| Host | Guest | Ka (M−1) | KS/KR | −ΔG0 (kJ mol−1) | ΔΔG0 (kJ mol−1) |
|---|---|---|---|---|---|
| (S,S,S)-94 | |||||
| (R)-98 | (2.0 ± 0.23) × 104 | 2.0 | 24.62 | 1.63 | |
| (S)-98 | (4.0 ± 0.34) × 104 | 26.25 | |||
| (R)-97 | (1.0 ± 0.42) × 104 | 5.0 | 22.82 | 3.99 | |
| (S)-97 | (5.0 ± 0.36) × 104 | 26.81 | |||
| (S,S,S)-95 | |||||
| (R)-98 | (2.0 ± 0.31) × 105 | 0.9 | 30.24 | −0.26 | |
| (S)-98 | (1.8 ± 0.38) × 105 | 29.98 | |||
| (R)-97 | (1.67 ± 0.19) × 104 | 2.4 | 24.09 | 2.16 | |
| (S)-97 | (4.0 ± 0.43) × 104 | 26.25 | |||
| (S,S,S)-96 | |||||
| (R)-98 | (1.5 ± 0.26) × 104 | 2.1 | 23.82 | 1.80 | |
| (S)-98 | (3.1 ± 0.18) × 104 | 25.62 | |||
| (R)-97 | (1.0 ± 0.34) × 104 | 5.0 | 22.82 | 3.99 | |
| (S)-97 | (5.0 ± 0.46) × 104 | 26.81 |
| Entry | Host | Guest a | Ka (M−1) b | KaS/KaRc | −ΔG0 (kJ mol−1) d | ΔΔG0 (kJ mol−1) e | ED f |
|---|---|---|---|---|---|---|---|
| 1 | (S,S)-102a | (R)-98 | 33.6 ± 0.3 | 3.86 | 8.71 | 3.34 | 59(S) |
| 2 | (S,S)-102a | (S)-98 | 129.6 ± 0.5 | 12.05 | |||
| 3 | (S,S)-102a | (R)-99 | 327.1 ± 0.2 | 31.0 | 14.35 | 8.50 | 94(S) |
| 4 | (S,S)-102a | (S)-99 | 10,138 ± 71 | 22.85 | |||
| 5 | (S,S)-102b | (R)-98 | 1852 ± 17 | 1.44 | 18.64 | 0.91 | 18(S) |
| 6 | (S,S)-102b | (S)-98 | 2668 ± 10 | 19.55 | |||
| 7 | (S,S)-102b | (R)-99 | 721.4 ± 3.1 | 0.36 | 16.31 | −2.51 | 47(R) |
| 8 | (S,S)-102b | (S)-99 | 262.6 ± 2.6 | 13.80 | |||
| 9 | (S,S)-102c | (R)-98 | 2559 ± 7 | 1.05 | 19.44 | 0.20 | 2(S) |
| 10 | (S,S)-102c | (S)-98 | 2681 ± 5 | 19.56 | |||
| 11 | (S,S)-102c | (R)-99 | 1126 ± 4 | 1.34 | 17.41 | 0.73 | 30(S) |
| 12 | (S,S)-102c | (S)-99 | 1511 ± 4 | 18.14 | |||
| 13 | (R,R)-103 | (R)-98 | 5412 ± 17 | 21.30 | |||
| 14 | (R,R)-103 | (S)-98 | nd g | nd g | |||
| 15 | (R,R)-103 | (R)-99 | 4719 ± 9 | 0.60 | 20.96 | −1.27 | 25(R) |
| 16 | (R,R)-103 | (S)-99 | 2833 ± 7 | 19.69 |
| Entry | Host | Guests | Kass (M−1) | KD/KL | −ΔG0 (kJ mol−1) a | −ΔΔG0 (kJ mol−1) b |
|---|---|---|---|---|---|---|
| 1 | 107 | d-PheOMe.HCl | 1785 | 2.04 | 18.3 | 1.80 |
| 2 | 107 | l-PheOMe.HCl | 875 | 16.5 | ||
| 3 | 107 | d-ValOMe.HCl | 2325 | 2.63 | 18.9 | 2.30 |
| 4 | 107 | l-ValOMe.HCl | 885 | 16.6 | ||
| 5 | 108 | d-PheOMe.HCl | 2580 | 1.22 | 1.2 | 0.5 |
| 6 | 108 | l-PheOMe.HCl | 2105 | 18.7 | ||
| 7 | 108 | d-ValOMe.HCl | 13,590 | 5.08 | 23.3 | 4.00 |
| 8 | 108 | l-ValOMe.HCl | 2675 | 19.3 | ||
| 9 | 109 | d-PheOMe.HCl | 395 | 0.77 (KL/KD = 1.29) | 14.6 | 0.60 |
| 10 | 109 | l-PheOMe.HCl | 510 | 15.2 | ||
| 11 | 109 | d-ValOMe.HCl | 32 | 0.33 (KL/KD = 3.00) | 8.5 | 2.80 |
| 12 | 109 | l-ValOMe.HCl | 96 | 11.3 | ||
| 13 | 110 | d-PheOMe.HCl | 1190 | 1.21 | 17.3 | 0.50 |
| 14 | 110 | l-PheOMe.HCl | 983 | 16.8 | ||
| 15 | 110 | d-ValOMe.HCl | 660 | 0.72 (KL/KD = 1.38) | 15.8 | 0.8 |
| 16 | 110 | l-ValOMe.HCl | 914 | 16.6 |
 | ΔΔδ | (ppm) | |||
| No. | Comp. No. | R1 | R2 | (S,S,S)-129 | (R,R,S)-130 |
| 1 | 131a | H | H | 0.03 | 0.74 |
| 2 | 131b | H | Ome | _b | 0.68 |
| 3 | 131c | H | i-Pr | _b | 0.76 |
| 4 | 131d | NO2 | H | _b | 0.81 |
| 5 | 131e | Br | H | _b | 0.40 |

| No. | Comp. No. | R | (S,S,S)-129 | (R,R,S)-130 |
|---|---|---|---|---|
| 1 | 132a | H | 0.17 | 0.04 |
| 2 | 132b | Me | 0.19 | _b |
| 3 | 133c | Cl | 0.16 | _b |
| 4 | 133 | - | 0.17 | _b |
| 5 | 134 | - | _b | _b |
| 6 | 135a | H | 0.40 | _b |
| 7 | 135b | Me | 0.42 | _b |
| 8 | 135c | Cl | 0.45 | 0.10 |
| 9 | 135d | OMe | 0.43 | _b |
| 10 | 135e | NO2 | 0.40 | 0.12 |
| 11 | 136 | - | 0.37 | _b |
| Entry | Host | Guests | Ka (M−1) | KD/KL | −ΔG0 (kJ mol−1) a | −ΔΔG0 (kJ mol−1) b |
|---|---|---|---|---|---|---|
| 1 | 148 | l-Phe-OMe | 89.4 | 1.12 | 11.13 | −0.27 |
| 2 | 148 | d-Phe-OMe | 99.8 | 11.40 | ||
| 3 | 149 | l-Ala-OMe | 437 | 1.45 | 15.06 | −0.93 |
| 4 | 149 | d-Ala-OMe | 634 | 15.99 | ||
| 5 | 149 | l-Val-OMe | 299 | 2.05 | 14.12 | −1.78 |
| 6 | 149 | d-Val-OMe | 613 | 15.90 | ||
| 7 | 149 | l-Leu-OMe | 260 | 2.24 | 13.78 | −2.00 |
| 8 | 149 | d-Leu-OMe | 583 | 15.78 | ||
| 9 | 149 | l-Phe-OMe | 319 | 3.33 | 14.28 | −2.99 |
| 10 | 149 | d-Phe-OMe | 1063 | 17.27 | ||
| 11 | 149 | l-Trp-OMe | 1238 | 2.68 | 17.64 | −2.44 |
| 12 | 149 | d-Trp-OMe | 3314 | 20.08 | ||
| 13 | 149 | l-Ala-OMe.HCl | 471 | 2.80 | 15.25 | −2.55 |
| 14 | 149 | d-Ala-OMe.HCl | 1319 | 17.80 | ||
| 15 | 149 | l-Leu-OMe.HCl | 327 | 3.52 | 14.35 | −3.11 |
| 16 | 149 | d-Leu-OMe.HCl | 1150 | 17.46 | ||
| 17 | 150 | l-Phe-OMe | 224 | 2.33 | 13.41 | −2.10 |
| 18 | 150 | d-Phe-OMe | 523 | 15.51 | ||
| 19 | 151 | l-Phe-OMe | 217 | 2.00 | 13.33 | −1.71 |
| 20 | 151 | d-Phe-OMe | 433 | 15.40 | ||
| 21 | 152 | l-Phe-OMe | 149 | 1.40 | 12.40 | −0.82 |
| 22 | 152 | d-Phe-OMe | 208 | 13.22 |
| Complex | Ka |
|---|---|
| 156a. N-Boc-l-Phe, -d-Phe | 100,65 |
| 156b. N-Boc-l-Ser, -d-Ser | <6065 |
| 156a. N-Boc-l-Phe, -d-Phe | 6565 |
| 156b. N-Boc-l-Ser, -d-Ser | 120,270 |
| Entry | Host a,b | Guest | K/dm3 mol | KL/KD | −ΔG0/KJ mol−1 | −ΔΔG0/KJ mol−1 |
|---|---|---|---|---|---|---|
| 1 | 158a2Br− | l-Phe | 10,328 | 1.29 | 23.05 | 0.63 |
| 2 | 158a2Br− | d-Phe | 8012 | 22.42 | ||
| 3 | 158a2Br− | l-Thr | 766 | 1.82 | 16.56 | 1.49 |
| 4 | 158a2Br− | d-Thr | 421 | 15.07 | ||
| 5 | 158b2Br− | l-Phe | 19,518 | 1.89 | 24.64 | 1.59 |
| 6 | 158b2Br− | d-Phe | 10,324 | 23.05 | ||
| 7 | 158b2Br− | l-His | 7627 | 2.03 | 22.30 | 1.77 |
| 8 | 158b2Br− | d-His | 3761 | 20.53 | ||
| 9 | 158c2Br− | l-Phe | 14,026 | 1.71 | 23.82 | 1.34 |
| 10 | 158b2Br− | d-Phe | 8222 | 22.48 | ||
| 11 | 159a2Br− | l-Ala | 820 | 1.61 | 16.73 | 1.19 |
| 12 | 159a2Br− | d-Ala | 509 | 15.54 | ||
| 13 | 159a2Br− | l-Thr | 1901 | 2.10 | 18.83 | 1.85 |
| 14 | 159a2Br− | d-Thr | 904 | 16.98 | ||
| 15 | 159a2Br− | l-Phe | 13,053 | 2.51 | 23.64 | 2.75 |
| 16 | 159a2Br− | d-Phe | 5196 | 20.89 | ||
| 17 | 159a2Br− | l-His | 11,639 | 3.03 | 23.35 | 2.76 |
| 18 | 159a2Br− | d-His | 3841 | 20.59 | ||
| 19 | 159a2Br− | l-Phe | 28,321 | 3.35 | 25.57 | 3.02 |
| 20 | 159a2Br− | d-Phe | 8458 | 22.55 | ||
| 21 | 159a2Br− | l-His | 12,031 | 3.20 | 23.43 | 2.91 |
| 22 | 159a2Br− | d-His | 3741 | 20.52 | ||
| 23 | 159c2Br− | l-Phe | 14,733 | 2.14 | 23.94 | 1.90 |
| 24 | 159c2Br− | d-Phe | 6899 | 22.04 | ||
| 25 | 158a2PF6− | BOC-l-His-Ome | 870 | 3.50 | 16.88 | 3.13 |
| 26 | 158a2PF6− | BOC-d-His-Ome | 248 | 13.75 | ||
| 27 | 158b2PF6− | BOC-l-His-Ome | 1001 | 4.07 | 17.23 | 3.50 |
| 28 | 158b2PF6− | BOC-d-His-Ome | 246 | 13.73 | ||
| 29 | 159a2PF6− | l-Ala-Ome | 610 | 1.72 | 16.00 | 1.50 |
| 30 | 159a2PF6− | d-Ala-OMe | 355 | 14.50 | ||
| 31 | 159a2PF6− | l-Phe-OMe | 9294 | 2.72 | 22.79 | 2.49 |
| 32 | 159a2PF6− | l-Phe-OMe | 3421 | 20.30 | ||
| 33 | 159a2PF6− | BOC-l-Phe-OMe | 9166 | 3.34 | 22.76 | 3.01 |
| 34 | 159a2PF6− | BOC-d-Phe-OMe | 2741 | 19.74 | ||
| 35 | 159b2PF6− | l-Phe-OMe | 10,720 | 3.14 | 23.15 | 2.86 |
| 36 | 159b2PF6− | d-Phe-OMe | 3412 | 20.29 | ||
| 37 | 159b2PF6− | BOC-l-Phe-OMe | 13,903 | 4.03 | 23.79 | 3.47 |
| 38 | 159b2PF6− | BOC-l-Phe-OMe | 3446 | 20.32 | ||
| 39 | 159b2PF6− | BOC-l-His-OMe | 1284 | 5.10 | 17.48 | 3.69 |
| 40 | 159b2PF6− | BOC-d-His-OMe | 252 | 13.79 |
| Entry | Host a,b | Guest c | K/dm3 mol | KL/KD | −ΔG0/Kj mol−1 | −ΔΔG0/Kj mol−1 d |
|---|---|---|---|---|---|---|
| 1 | 158a | l-Ala | 1623 | 1.81 | 18.44 | 1.49 |
| d-Ala | 897 | 16.95 | ||||
| 2 | 158a | l-Phe | 33,748 | 1.75 | 26.01 | 1.41 |
| d-Phe | 19,241 | 24.60 | ||||
| 3 | 158a | l-His | 7458 | 1.92 | 22.24 | 1.63 |
| d-His | 3876 | 20.61 | ||||
| 4 | 159a | l-Phe-OMe | 4099 | 1.59 | 20.99 | 1.16 |
| d-Phe-OMe | 2515 | 19.83 | ||||
| 5 | 158b | l-Ala | 1682 | 1.82 | 18.53 | 1.51 |
| d-Ala | 921 | 17.02 | ||||
| 6 | 158b | l-Phe | 68,274 | 3.36 | 27.76 | 3.02 |
| d-Phe | 20,310 | 24.74 | ||||
| 7 | 158b | l-His | 9510 | 3.04 | 22.85 | 2.78 |
| d-His | 3129 | 20.07 | ||||
| 8 | 159b | l-Ala-OMe | 1704 | 1.95 | 18.55 | 1.86 |
| d-Ala-OMe | 874 | 16.69 | ||||
| 9 | 159b | BOC-l-Ala-OMe | 1453 | 2.10 | 18.16 | 1.86 |
| BOC-d-Ala-OMe | 690 | 16.30 | ||||
| 10 | 159b | l-Phe-OMe | 6882 | 2.22 | 22.04 | 1.98 |
| d-Phe-OMe | 3105 | 20.06 | ||||
| 11 | 159b | BOC-l-Phe-OMe | 6736 | 2.68 | 21.99 | 2.47 |
| BOC-d-Phe-OMe | 2510 | 19.52 | ||||
| 12 | 159b | BOC-l-His-OMe | 2941 | 4.10 | 19.92 | 3.52 |
| BOC-d-His-OMe | 717 | 16.40 |
| Amino Acid a | (ΔI/I0)max | Ef |
|---|---|---|
| Ala | 1.50 | 1.52 |
| Phe | 1.58 | 1.68 |
| Pro | 1.44 | 1.61 |
| Ser | 2.60 | 4.04 |
| Met | 1.44 | 1.33 |
| Amino Acids | [197-Hg2+] | [199-Hg2+] | ||||
|---|---|---|---|---|---|---|
| KL (105 M−1) | KD (105 M−1) | KL/KD | KL (105 M−1) | KD (105 M−1) | KL/KD | |
| Ala | 1.284 | 1.252 | 1.026 | 0.4664 | 0.4646 | 1.004 |
| Val | 1.226 | 1.199 | 1.023 | 0.4358 | 0.4352 | 1.001 |
| His | 2.201 | 2.002 | 1.100 | 0.8610 | 0.8455 | 1.018 |
| Cys | 2.510 | 2.121 | 1.183 | 1.630 | 1.616 | 1.008 |
| Met | 2.032 | 1.841 | 1.104 | 0.7667 | 0.7764 | 0.9875 |
| Guests | First CE | Second CE | Δ ε(cm−1 M−1) | g = Δ ε/ε × 10−3 (at First CE) | Binding Mode |
|---|---|---|---|---|---|
| (R)-206 | − | + | 498.65 | 35.9 | A |
| (R)-207 | − | + | 121.10 | 15.16 | A |
| (R)-208 | − | + | 85.48 | 10.8 | A |
| (R)-209 | − | + | 7.12 | 10.0 | A |
| (R)-210 | + | − | 124.66 | 10.5 | B |
| (R)-211 | + | − | 641.13 | 9.3 | B |
| (1R,2S)-212 | + | − | 4.63 | 0.1 | B |
| (1R,2S)-213 | + | − | 3.21 | 0.4 | B |
| Racemic Ammonium Guest | Enantioselectivity a | Extraction (%) b |
|---|---|---|
| α-phenylethylamine | 71(R):29(S) | 82 |
| α-(1-naphthyl)ethylamine | 70:30 | 99 |
| Phenylglycine methyl ester | 78(S):22(R) | 60 |
| Tryptophan methyl ester | 67(S):33(R) | 57 c |
| Alanine methyl ester | 53(S):47(R) | 41 |
| Phenylalanine methyl ester | 55(S):45(R) | 36 |

| Entry | Racemic Guest | Enantioselectivity a | Extraction (%) b |
|---|---|---|---|
| 1 | 216 | 63:37 c | 50 |
| 2 | 217 | 75:25 | 60 |
| 3 | 218 | 72:28 | 40 |
| 4 | 219 | 50:50 | 97 |
| 5 | 220 | 58:42 | 72 |
| 6 | 221 | 58:42 | 71 |
| 7 | 222 | 71:29 | <5 |
| 8 | 223 | 61:39 | 10 |
| 9 | 224 | 83:17 | <5 |

| Entry | Racemic Guest | Enantioselectivity | Extraction (%) |
|---|---|---|---|
| 1 | 228 | 66:34 c | 74 |
| 2 | 229 | 61:39 | 20 |
| 3 | 230 | 56:44 | 26 |
| 4 | 231 | 53:37 c | 41 |
| 5 | 232 | 55:45 c | 36 |
| Entry | Receptor | Ammonium Guest | Enantioselectivity | Extraction (%) |
|---|---|---|---|---|
| 1 | 235 | PhCH(NH3+) CH3 | 71(R):29(S) | 82 |
| 2 | 235 | PhCH(NH3+) CO2Me | 78(S):22(R) | 60 |
| 3 | 234 | PhCH(NH3+) CH3 | 64(R):36(S) | 51 |
| 4 | 234 | PhCH(NH3+) CO2Me | 70(S):30(R) | 22 |
| 5 | 236a | PhCH(NH3+) CH3 | 59(S):41(R) | 100 |
| 6 | 236a | PhCH(NH3+) CO2Me | 62(S):38(R) | 91 |
| 7 | 236b | PhCH(NH3+) CH3 | 58(S):42(R) | 80 |
| 8 | 236b | PhCH(NH3+) CO2Me | 69(S):31(R) | 69 |
| 9 | 236c | PhCH(NH3+) CH3 | 50(S):50(R) | - |
| 10 | 237 | PhCH(NH3+) CH3 | 50(S):50(R) | 70 |
| Receptor-Guest | Temp (°C) | Enantioselectivity | Binding (%) a |
|---|---|---|---|
| 235-(R,S)-98 | 25 | 71(R):29(S) c | 82 |
| 240a-(R,S)-98 | −30 | 57(R):43(S) | ~100 |
| −50 | 60(R):40(S) | ~100 | |
| 240b-(R,S)-98 | −30 | ||
| −50 | 61(R):39(S) | 73 | |
| 235-(R,S)-Ala Methyl ester | 25 | 47(R):53(S) c | 41 |
| 240a-(R,S)-Ala Methyl ester | 10 | 61(R):39(S) | 62 |
| −10 | 60(R):40(S) | 65 | |
| −30 | 61(R):39(S) | 67 | |
| −50 | 64(R):36(S) | 68 | |
| 240b-(R,S)-Ala Methyl ester | −10 | 64(R):36(S) | 79 |
| −30 | 66(R):34(S) | 83 | |
| −50 | 72(R):28(S) | 76 |
| Guest | Ka |
|---|---|
| Ethoxycarbonyl-l-proline | 57.0 |
| Cbz-l-phenylglycine | 16.0 |
| Cbz-l-phenylalanine | 15.0 |
| Ethoxycarbonyl-l-alanine | 8.4 |
| Ethoxycarbonyl-l-leucine | 7.6 |
| BOC-l-leucine | 4.0 |
| Chiral Selectors | N-Ac-amino acid Tetrabutyl Ammonium Salt | Association Constants (M−1) | Discrimination |
|---|---|---|---|
 | (l)-N-Ac-Phe-COO– (d)-N-Ac-Phe-COO– (l)-N-Ac-Val-COO– (d)-N-Ac-Val-COO– | 540 330 600 700 | L/D 1.60 D/L 1.15 a |
 | (l)-N-Ac-Phe-COO– (d)-N-Ac-Phe-COO– | 1150 2300 | D/L 2.00 |
 | (l)-N-Ac-Phe-COO– (d)-N-Ac-Phe-COO– | 800 960 | D/L 1.20 a |
 | (l)-N-Ac-Phe-COO– (d)-N-Ac-Phe-COO– (l)-N-Ac-Val-COO– (d)-N-Ac-Val-COO– | 1250 1550 2150 2350 | L/D 1.25 a D/L 1.10 a |
| (aR) 247i (aS) 247i | (l)-N-Ac-Leu-COO- | 2750 3900 | 1.42 |
| (aR) 247i (aS) 247i | (S)-Naproxenate | 1330 1900 | 1.43 |
| (aR) 247i (aS) 247i | (l)-N-Ac-Trp-COO- | 825 1150 | 1.47 |
| Guests | Observed Signals | Δδ(ppm) 248 249 | ΔΔ δ (Hz) 248 249 | ||
|---|---|---|---|---|---|
 | –CH | −0.13 −0.12 −0.205 −0.217 | −0.122 −0.14 - | 4 4.8b | 7.2 - |
 | –CH | −0.08 −0.11 | - | 12 | 0 |
 | Ortho CH of phenyl ring | −0.060 −0.008 | −0.101 −0.137 | 21.2 | 14.4c |
 | Ortho CH of toluoyl ring | −0.001 −0.050 | −0.05 0.102 | 20.8 | 20.4c |
| Entry | Guest | 218 | 219 | 220 |
|---|---|---|---|---|
| 1 | (R)-PEA | <1 | 200(± 30) | 30000(± 11000) b |
| 2 | (S)-PEA | <1 | 480(± 60) | 4500(± 590) |
| Guest | Ks | Δ δmax | Selectivity Coefficients | −ΔG0 kJ mol−1 | ΔΔG0 kJ mol−1 |
|---|---|---|---|---|---|
| 252*(R)-PEA | 200(± 40) | 0.03 | 13.1 | ||
| 252*(S)-PEA | 480(± 70) | 0.06 | 2.4 | 15.3 | −2.2 |
| 252*(R)-PAM | 16,000(± 4900) | 0.01 | 24 | ||
| 252*(S)-PAM | 1900(± 500) | 0.02 | 8.4 | 18.7 | 5.3 |
| 252*(R)-BA | 130(± 40) | 0.01 | 12.1 | ||
| 252*(S)-BA | 940(± 240) | 0.01 | 7.2 | 17 | −4.9 |
| 252*(R)-NEA | _ | −d | - | ||
| 252*(S)-NEA | d | −d | - | - | - |
| 252*(R)-BEA | 560(± 210) | 0.01 | 15.7 | ||
| 252*(S)-BEA | 540(± 50) | 0.05 | 1.0 | 15.6 | 0.1 |
| 252*(R)-AH | 360(± 70) | 0.01 | 14.6 | ||
| 252*(S)-AH | 100(± 20) | 0.02 | 3.6 | 11.4 | 3.2 |
| 253*(R)-PEA | 30,000(± 11,000) | 0.25 | 25.5 | ||
| 253*(S)-PEA | 4500(± 590) | 0.30 | 6.7 | 20.8 | 4.7 |
| 253*(R)-PAM | 2000(± 240) | 0.22 | 18.8 | ||
| 253*(S)-PAM | 1100(± 270) | 0.22 | 1.8 | 17.4 | 1.5 |
| 253*(R)-BA | 1600(± 260) | 0.17 | 18.3 | ||
| 253*(S)-BA | 2400(± 930) | 0.20 | 1.5 | 19.3 | −1.0 |
| 253*(R)-NEA | 1000(± 180) | 0.06 | 17.1 | ||
| 253*(S)-NEA | 610(± 110) | 0.06 | 1.6 | 15.9 | 1.2 |
| 253*(R)-BEA | 6600(± 1600) | 0.30 | 21.8 | ||
| 253*(S)-BEA | 3200(± 450) | 0.28 | 2.1 | 20 | 1.8 |
| 253*(R)-AH | 9700(± 3100) | 0.13 | 22.7 | ||
| 253*(S)-AH | 2400(± 710) | 0.16 | 4.0 | 19.3 | 3.5 |

| Entry | Substrate | R/S a |
|---|---|---|
| 1 |  | 18:1 b |
| 2 |  | 23:1 c |
| 3 |  | 7:1 d |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khose, V.N.; John, M.E.; Pandey, A.D.; Borovkov, V.; Karnik, A.V. Chiral Heterocycle-Based Receptors for Enantioselective Recognition. Symmetry 2018, 10, 34. https://doi.org/10.3390/sym10020034
Khose VN, John ME, Pandey AD, Borovkov V, Karnik AV. Chiral Heterocycle-Based Receptors for Enantioselective Recognition. Symmetry. 2018; 10(2):34. https://doi.org/10.3390/sym10020034
Chicago/Turabian StyleKhose, Vaibhav N., Marina E. John, Anita D. Pandey, Victor Borovkov, and Anil V. Karnik. 2018. "Chiral Heterocycle-Based Receptors for Enantioselective Recognition" Symmetry 10, no. 2: 34. https://doi.org/10.3390/sym10020034
APA StyleKhose, V. N., John, M. E., Pandey, A. D., Borovkov, V., & Karnik, A. V. (2018). Chiral Heterocycle-Based Receptors for Enantioselective Recognition. Symmetry, 10(2), 34. https://doi.org/10.3390/sym10020034