Free Radicals Mediated Redox Signaling in Plant Stress Tolerance
Abstract
:1. Introduction
2. Chemical Biology of RSS in Plants
3. RSS Biosynthesis in Plants
4. RSS Signaling in Plants
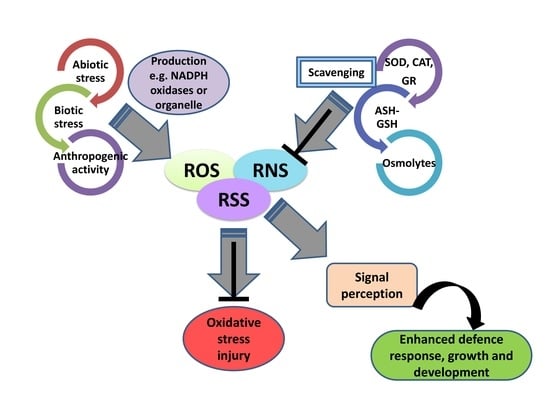
5. Crosstalk between ROS/RNS/RSS in Plant Defense
| Processes | Functional Mechanisms | Stress Conditions | Pathway Involved | References |
|---|---|---|---|---|
| Seed germination | ||||
| H2O2 | Break seed dormancy and promote seed germination by initiating protein carbonylation | Drought, heat, and salinity | GA signaling pathway | [44,45] |
| NO | Regulate seed germination and pollen tube growth by activating catabolic enzymes of ABA and GA biosynthesis | Osmotic and heavy metals | ABA and GA signaling pathways | [46] |
| H2S | Promote seed germination of wheat plants by enhancing amylase and esterase activities | Heavy metal and salt | Antioxidant defense pathway | [47,48] |
| Stomatal movement | ||||
| H2O2 | Induce stomatal closure by regulating the activity of NADPH oxidase and control the level of ROS modulates’ stomatal opening by invoking plant peroxisome—specific autophagy | Drought and pathogen attack | ABA and antioxidant signaling pathways | [49,50] |
| NO | Promote stomatal closure by inducing tyrosine nitration and S-nitrosation of ABA receptors | Drought and salinity | ABA and antioxidant signaling pathways | [51,52] |
| H2S | Induce stomatal closure by affecting the activities of ion channels via inducing sulfhydration | Drought and cold | ABA and MAPK signaling | [51,53] |
| Root organogenesis | ||||
| H2O2 | Instigate de novo root organogenesis by acting downstream of auxin and Ca2+ signaling | Oxidative stress | Auxin signaling pathway | [54,55] |
| NO | Modulate root organogenesis by activating the expression of MYB and BHLH transcription factors | Oxidative stress | Auxin and jasmonic acid signaling pathways | [54] |
| H2S | Induce adventitious root formation by acting as upstream of IAA and NO signaling | Heat and heavy metal | Auxin and abscisic acid signaling pathways | [56,57] |
| Leaf senescence /fruit ripening | ||||
| H2O2 | Delay leaf senescence/fruit ripening by regulating the ascorbate-glutathione cycle | Oxidative stress | Antioxidant defense pathway | [58] |
| NO | Delay induced leaf senescence/fruit ripening by enhancing the expression of stress-responsive genes, ACC synthase, and oxidase enzymes | Salt stress | Antioxidant and ABA signaling pathways | [59,60] |
| H2S | Delay leaf senescence/fruit ripening by regulating the expression of the Des1 gene | Oxidative stress | Abscisic acid signaling pathways | [56,61] |
| Post-translational/ epigenetic regulation | ||||
| H2O2 | Induce oxidative posttranslational modifications, DNA methylation, and histone modification, thus stimulating plant stress response | Abiotic/biotic stress | Stress defense pathway | [62,63] |
| NO | Induce tyrosine nitration and S-nitrosation and epigenetic regulation of various small and long non-coding RNAs, thereby regulating plant immune system | Abiotic/biotic stress | Stress defense pathway | [63,64] |
| H2S | Induce S-Sulfhydration of cysteine residues, thereby activating plant tolerance | Abiotic/biotic stress | Stress defense pathway | [65] |
5.1. Regulation of Gene Expression
5.2. Regulation of Post-Translational Modification
5.3. Regulation of Epigenetic Modification
6. Potential Biotechnological Application in Agriculture
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Talbi, S.; Romero-Puertas, M.C.; Hernández, A.; Terrón, L.; Ferchichi, A.; Sandalio, L.M. Drought tolerance in a Saharian plant Oudneya africana: Role of antioxidant defences. Environ. Exp. Bot. 2015, 111, 114–126. [Google Scholar] [CrossRef]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 2017, 15, 207–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blum, A. Osmotic adjustment is a prime drought stress adaptive engine in support of plant production. Plant Cell Environ. 2017, 40, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. ROS are good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cejudo, F.J.; Sandalio, L.M.; Van Breusegem, F. Understanding plant responses to stress conditions: Redox-based strategies. J. Exp. Bot. 2021, 72, 5785–5788. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; González-Gordo, S.; Palma, J.M. Nitric oxide and hydrogen sulfide modulate the NADPH-generating enzymatic system in higher plants. J. Exp. Bot. 2021, 72, 830–847. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Suzuki, N.; Miller, G.A.D.; Tognetti, V.B.; Vandepoele, K.; Gollery, M.; Shulaev, V.; Van Breusegem, F. ROS signaling: The new wave? Trends Plant Sci. 2011, 16, 300–309. [Google Scholar] [CrossRef]
- Del Río, L.A. ROS and RNS in plant physiology: An overview. J. Exp. Bot. 2015, 66, 2827–2837. [Google Scholar] [CrossRef] [Green Version]
- Gruhlke, M.C.; Slusarenko, A.J. The biology of reactive sulfur species (RSS). Plant Physiol. Biochem. 2012, 59, 98–107. [Google Scholar] [CrossRef]
- Corpas, F.J.; Barroso, J.B. Reactive sulfur species (RSS): Possible new players in the oxidative metabolism of plant peroxisomes. Front. Plant Sci. 2015, 6, 116. [Google Scholar] [CrossRef]
- Corpas, F.J.; Barroso, J.B.; González-Gordo, S.; Muñoz-Vargas, M.A.; Palma, J.M. Hydrogen sulfide: A novel component in Arabidopsis peroxisomes which triggers catalase inhibition. J. Integr. Plant Biol. 2019, 61, 871–883. [Google Scholar] [CrossRef] [Green Version]
- Aroca, A.; Gotor, C.; Romero, L.C. Hydrogen sulfide signaling in plants: Emerging roles of protein persulfidation. Front. Plant Sci. 2018, 9, 1369. [Google Scholar] [CrossRef] [Green Version]
- Corpas, F.J.; González-Gordo, S.; Palma, J.M. Plant peroxisomes: A factory of reactive species. Front. Plant Sci. 2020, 11, 853. [Google Scholar] [CrossRef]
- Lau, N.; Pluth, M.D. Reactive sulfur species (RSS): Persulfides, polysulfides, potential, and problems. Curr. Opin. Chem. Biol. 2019, 49, 1–8. [Google Scholar] [CrossRef]
- Mishanina, T.V.; Libiad, M.; Banerjee, R. Biogenesis of reactive sulfur species for signaling by hydrogen sulfide oxidation pathways. Nat. Chem. Biol. 2015, 11, 457–464. [Google Scholar] [CrossRef] [Green Version]
- Giles, G.I.; Jacob, C. Reactive sulfur species: An emerging concept in oxidative stress. Biol. Chem. 2002, 383, 375–388. [Google Scholar] [CrossRef]
- Wächtershäuser, G. From volcanic origins of chemoautotrophic life to Bacteria, Archaea and Eukarya. Philos. Trans. R. Soc. B. 2006, 361, 1787–1808. [Google Scholar] [CrossRef] [Green Version]
- Zhou, M.; Zhang, J.; Zhou, H.; Zhao, D.; Duan, T.; Wang, S.; Yuan, X.; Xie, Y. Hydrogen sulfide-linked persulfidation maintains protein stability of Abscisic Acid-Insensitive 4 and delays seed germination. Int. J. Mol. Sci. 2022, 23, 1389. [Google Scholar] [CrossRef]
- Giles, G.I.; Tasker, K.M.; Jacob, C. Hypothesis: The role of reactive sulfur species in oxidative stress. Free Radic. Biol. Med. 2001, 31, 1279–1283. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, A.; Galvan, A.; Fernandez, E. Nitrate reductase regulates plant nitric oxide homeostasis. Trends Plant Sci. 2017, 22, 163–174. [Google Scholar] [CrossRef]
- Jacob, C.; Anwar, A. The chemistry behind redox regulation with a focus on sulphur redox systems. Physiol. Plant. 2008, 133, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Gruhlke, M.C. Reactive sulfur species: A new player in plant physiology. In Reactive Oxygen, Nitrogen and Sulfur Species in Plants: Production, Metabolism, Signaling and Defense Mechanisms; Hasanuzzaman, M., Fotopoulos, V., Nahar, K., Fujita, M., Eds.; Wiley: Hoboken, NJ, USA, 2019; pp. 715–728. [Google Scholar] [CrossRef]
- Giles, G.I.; Nasim, M.J.; Ali, W.; Jacob, C. The reactive sulfur species concept: 15 years on. Antioxidants 2017, 6, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olson, K.R. Reactive oxygen species or reactive sulfur species: Why we should consider the latter. J. Exp. Biol. 2020, 223, 196352. [Google Scholar] [CrossRef] [PubMed]
- Olson, K.R.; Gao, Y.; Arif, F.; Arora, K.; Patel, S.; DeLeon, E.R.; Sutton, T.R.; Feelisch, M.; Cortese-Krott, M.M.; Straub, K.D. Metabolism of hydrogen sulfide (H2S) and production of reactive sulfur species (RSS) by superoxide dismutase. Redox Biol. 2018, 15, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.; Reumann, S.; Lisik, P.; Tietz, S.; Olsen, L.J.; Hu, J. Proteome analysis of peroxisomes from dark-treated senescent Arabidopsis leaves. J. Integr. Plant Biol. 2018, 60, 1028–1050. [Google Scholar] [CrossRef] [Green Version]
- Zaffagnini, M.; Fermani, S.; Marchand, C.H.; Costa, A.; Sparla, F.; Rouhier, N.; Geigenberger, P.; Lemaire, S.D.; Trost, P. Redox homeostasis in photosynthetic organisms: Novel and established thiol-based molecular mechanisms. Antioxid. Redox Signal. 2019, 31, 155–210. [Google Scholar] [CrossRef]
- Biswas, S.; Mondal, R.; Srivastava, A.; Trivedi, M.; Singh, S.K.; Mishra, Y. In silico characterization, molecular phylogeny, and expression profiling of genes encoding legume lectin-like proteins under various abiotic stresses in Arabidopsis thaliana. BMC Genom. 2022, 23, 480. [Google Scholar] [CrossRef]
- Palma, J.M.; Mateos, R.M.; López-Jaramillo, J.; Rodríguez-Ruiz, M.; González-Gordo, S.; Lechuga-Sancho, A.M.; Corpas, F.J. Plant catalases as NO and H2S targets. Redox Biol. 2020, 24, 131–137. [Google Scholar] [CrossRef]
- Filipovic, M.R.; Zivanovic, J.; Alvarez, B.; Banerjee, R. Chemical biology of H2S signaling through persulfidation. Chem. Rev. 2018, 118, 1253–1337. [Google Scholar] [CrossRef]
- Hänsch, R.; Mendel, R.R. Sulfite oxidation in plant peroxisomes. Photosynth. Res. 2015, 86, 337–343. [Google Scholar] [CrossRef]
- Huang, H.; Ullah, F.; Zhou, D.X.; Yi, M.; Zhao, Y. Mechanisms of ROS regulation of plant development and stress responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef]
- Olson, K.R. Hydrogen sulfide, reactive sulfur species and coping with reactive oxygen species. Free Radic. Biol. Med. 2019, 140, 74–83. [Google Scholar] [CrossRef]
- Olson, K.R.; Gao, Y.; DeLeon, E.R.; Arif, M.; Arif, F.; Arora, N.; Straub, K.D. Catalase as a sulfide-sulfur oxido-reductase: An ancient (and modern?) regulator of reactive sulfur species (RSS). Redox Biol. 2017, 12, 325–339. [Google Scholar] [CrossRef]
- Pan, R.; Liu, J.; Wang, S.; Hu, J. Peroxisomes: Versatile organelles with diverse roles in plants. New Phytol. 2020, 225, 1410–1427. [Google Scholar] [CrossRef] [Green Version]
- Mangal, V.; Lal, M.K.; Tiwari, R.K.; Altaf, M.A.; Sood, S.; Kumar, D.; Bharadwaj, V.; Singh, B.; Singh, R.K.; Aftab, T. Molecular Insights into the Role of Reactive Oxygen, Nitrogen and Sulphur Species in Conferring Salinity Stress Tolerance in Plants. J. Plant Growth Regul. 2022, 1–21. [Google Scholar] [CrossRef]
- Fuentes-Lara, L.O.; Medrano-Macías, J.; Pérez-Labrada, F.; Rivas-Martínez, E.N.; García-Enciso, E.L.; González-Morales, S.; Juárez-Maldonado, A.; Rincón-Sánchez, F.; Benavides-Mendoza, A. From elemental sulfur to hydrogen sulfide in agricultural soils and plants. Molecules 2019, 24, 2282. [Google Scholar] [CrossRef] [Green Version]
- Shankar, S.; Jaiswal, L.; Rhim, J.W. New insight into sulfur nanoparticles: Synthesis and applications. Crit. Rev. Environ. Sci. Technol. 2021, 51, 2329–2356. [Google Scholar] [CrossRef]
- Corpas, F.J.; Palma, J.M. H2S signaling in plants and applications in agriculture. J. Adv. Res. 2020, 24, 131–137. [Google Scholar] [CrossRef]
- Rai, K.K.; Rai, N.; Rai, S.P. Salicylic acid and nitric oxide alleviate high temperature induced oxidative damage in Lablab purpureus L plants by regulating bio-physical processes and DNA methylation. Plant Physiol. Biochem. 2018, 128, 72–88. [Google Scholar] [CrossRef]
- Rai, K.K.; Rai, N.; Aamir, M.; Tripathi, D.; Rai, S.P. Interactive role of salicylic acid and nitric oxide on transcriptional reprogramming for high temperature tolerance in Lablab purpureus L.: Structural and functional insights using computational approaches. J. Biotechnol. 2020, 309, 113–130. [Google Scholar] [CrossRef] [PubMed]
- Rai, K.K.; Pandey, N.; Rai, S.P. Salicylic acid and nitric oxide signaling in plant heat stress. Physiol. Plant. 2020, 168, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Vivancos, P.; Barba-Espín, G.; Hernández, J.A. Elucidating hormonal/ROS networks during seed germination: Insights and perspectives. Plant Cell Rep. 2013, 32, 1491–1502. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C. The signalling role of ROS in the regulation of seed germination and dormancy. Biochem. J. 2019, 476, 3019–3032. [Google Scholar] [CrossRef] [PubMed]
- Arc, E.; Sechet, J.; Corbineau, F.; Rajjou, L.; Marion-Poll, A. ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front. Plant Sci. 2013, 4, 63. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Hu, L.Y.; Hu, K.D.; He, Y.D.; Wang, S.H.; Luo, J.P. Hydrogen sulfide promotes wheat seed germination and alleviates oxidative damage against copper stress. J. Integr. Plant Biol. 2008, 50, 1518–1529. [Google Scholar] [CrossRef]
- da-Silva, C.J.; Modolo, L.V. Hydrogen sulfide: A new endogenous player in an old mechanism of plant tolerance to high salinity. Acta Bot Brasilica. 2017, 32, 150–160. [Google Scholar] [CrossRef] [Green Version]
- Qi, J.; Song, C.P.; Wang, B.; Zhou, J.; Kangasjärvi, J.; Zhu, J.K.; Gong, Z. Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Biol. 2018, 60, 805–826. [Google Scholar] [CrossRef] [Green Version]
- Medeiros, D.B.; Barros, J.A.; Fernie, A.R.; Araújo, W.L. Eating away at ROS to regulate stomatal opening. Trends Plant Sci. 2020, 25, 220–223. [Google Scholar] [CrossRef]
- Mishra, V.; Singh, P.; Tripathi, D.K.; Corpas, F.J.; Singh, V.P. Nitric oxide and hydrogen sulfide: An indispensable combination for plant functioning. Trends Plant Sci. 2021, 26, 1270–1285. [Google Scholar] [CrossRef]
- Sun, L.R.; Yue, C.M.; Hao, F.S. Update on roles of nitric oxide in regulating stomatal closure. Plant Signal. Behav. 2019, 14, e1649569. [Google Scholar] [CrossRef]
- Jin, Z.; Wang, Z.; Ma, Q.; Sun, L.; Zhang, L.; Liu, Z.; Liu, D.; Hao, X.; Pei, Y. Hydrogen sulfide mediates ion fluxes inducing stomatal closure in response to drought stress in Arabidopsis thaliana. Plant Soil 2017, 419, 141–152. [Google Scholar] [CrossRef]
- Raya-González, J.; López-Bucio, J.S.; López-Bucio, J. Nitric oxide and hydrogen peroxide in root organogenesis. In Nitric Oxide and Hydrogen Peroxide Signaling in Higher Plants; Gupta, D., Palma, J., Corpas, F., Eds.; Springer: Cham, Switzerland, 2019. [Google Scholar] [CrossRef]
- Shin, S.Y.; Park, S.J.; Kim, H.S.; Jeon, J.H.; Lee, H.J. Wound-induced signals regulate root organogenesis in Arabidopsis explants. BMC Plant Biol. 2022, 22, 133. [Google Scholar] [CrossRef]
- Jin, Z.; Pei, Y. Physiological implications of hydrogen sulfide in plants: Pleasant exploration behind its unpleasant odour. Oxid. Med. Cell. Longev. 2015, 2015, 397502. [Google Scholar] [CrossRef] [Green Version]
- Arif, Y.; Hayat, S.; Yusuf, M.; Bajguz, A. Hydrogen sulfide: A versatile gaseous molecule in plants. Plant Physiol. Biochem. 2021, 158, 372–384. [Google Scholar] [CrossRef]
- Tan, X.L.; Zhao, Y.T.; Shan, W.; Kuang, J.F.; Lu, W.J.; Su, X.G.; Tao, N.G.; Lakshmanan, P.; Chen, J.Y. Melatonin delays leaf senescence of post-harvest Chinese flowering cabbage through ROS homeostasis. Food Res. Int. 2020, 138, 109790. [Google Scholar] [CrossRef]
- Kong, X.; Wang, T.; Li, W.; Tang, W.; Zhang, D.; Dong, H. Exogenous nitric oxide delays salt-induced leaf senescence in cotton (Gossypium hirsutum L.). Acta Physiol. Plant. 2016, 38, 61. [Google Scholar] [CrossRef]
- Bruand, C.; Meilhoc, E. Nitric oxide in plants: Pro-or anti-senescence. J. Exp. Bot. 2019, 70, 4419–4427. [Google Scholar] [CrossRef]
- Decros, G.; Baldet, P.; Beauvoit, B.; Stevens, R.; Flandin, A.; Colombié, S.; Gibon, Y.; Pétriacq, P. Get the balance right: ROS homeostasis and redox signalling in fruit. Front. Plant Sci. 2019, 10, 1091. [Google Scholar] [CrossRef] [Green Version]
- Waszczak, C.; Akter, S.; Jacques, S.; Huang, J.; Messens, J.; Van Breusegem, F. Oxidative post-translational modifications of cysteine residues in plant signal transduction. J. Exp. Bot. 2015, 66, 2923–2934. [Google Scholar] [CrossRef] [Green Version]
- Astier, J.; Lindermayr, C. Nitric oxide-dependent post-translational modification in plants: An update. Int. J. Mol. Sci. 2012, 13, 15193–15208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, K.J.; Kolbert, Z.; Durner, J.; Lindermayr, C.; Corpas, F.J.; Brouquisse, R.; Barroso, J.B.; Umbreen, S.; Palma, J.M.; Hancock, J.T.; et al. Regulating the regulator: Nitric oxide control of post-translational modifications. New Phytol. 2020, 227, 1319–1325. [Google Scholar] [CrossRef] [PubMed]
- Aroca, Á.; Serna, A.; Gotor, C.; Romero, L.C. S-sulfhydration: A cysteine post-translational modification in plant systems. Plant Physiol. 2015, 168, 334–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Signorelli, S.; Considine, M.J. Nitric oxide enables germination by a four-pronged attack on ABA-induced seed dormancy. Front. Plant Sci. 2018, 9, 296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prakash, V.; Singh, V.P.; Tripathi, D.K.; Sharma, S.; Corpas, F.J. Crosstalk between nitric oxide (NO) and abscisic acid (ABA) signalling molecules in higher plants. Environ. Exp. Bot. 2019, 161, 41–49. [Google Scholar] [CrossRef]
- Chen, S.; Jia, H.; Wang, X.; Shi, C.; Wang, X.; Ma, P.; Wang, J.; Ren, M.; Li, J. Hydrogen sulfide positively regulates abscisic acid signaling through persulfidation of SnRK2. 6 in guard cells. Mol. Plant 2020, 13, 732–744. [Google Scholar] [CrossRef]
- Liu, H.; Wang, C.; Li, C.; Zhao, Z.; Wei, L.; Liu, Z.; Hu, D.; Liao, W.; Bozhkov, P. Nitric oxide is involved in hydrogen sulfide-induced adventitious rooting in tomato (Solanum lycopersicum). Funct. Plant Biol. 2022, 49, 245–258. [Google Scholar] [CrossRef]
- Fang, H.; Liu, R.; Yu, Z.; Wu, G.; Pei, Y. Gasotransmitter H2S accelerates seed germination via activating AOX mediated cyanide-resistant respiration pathway. bioRxiv 2021. [Google Scholar] [CrossRef]
- Kasprowicz, A.; Szuba, A.; Volkmann, D.; Baluška, F.; Wojtaszek, P. Nitric oxide modulates dynamic actin cytoskeleton and vesicle trafficking in a cell type-specific manner in root apices. J. Exp. Bot. 2009, 60, 1605–1617. [Google Scholar] [CrossRef] [Green Version]
- Plohovska, S.H.; Krasylenko, Y.A.; Yemets, A.I. Nitric oxide modulates actin filament organization in Arabidopsis thaliana primary root cells at low temperatures. Cell Biol. Int. 2019, 43, 1020–1030. [Google Scholar] [CrossRef]
- Li, Z.G.; Long, W.B.; Yang, S.Z.; Wang, Y.C.; Tang, J.H. Signaling molecule methylglyoxal-induced thermotolerance is partly mediated by hydrogen sulfide in maize (Zea mays L.) seedlings. Acta Physiol. Plant. 2018, 40, 76. [Google Scholar] [CrossRef]
- Li, F.C.; Wang, J.; Wu, M.M.; Fan, C.M.; Li, X.; He, J.M. Mitogen-activated protein kinase phosphatases affect UV-B-induced stomatal closure via controlling NO in guard cells. Plant Physiol. 2017, 173, 760–770. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Du, Y.; Hou, Y.J.; Zhao, Y.; Hsu, C.C.; Yuan, F.; Zhu, X.; Tao, W.A.; Song, C.P.; Zhu, J.K. Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc. Nat. Acad. Sci. USA 2015, 112, 613–618. [Google Scholar] [CrossRef] [Green Version]
- Honda, K.; Yamada, N.; Yoshida, R.; Ihara, H.; Sawa, T.; Akaike, T.; Iwai, S. 8-Mercapto-cyclic GMP mediates hydrogen sulfide-induced stomatal closure in Arabidopsis. Plant Cell Physiol. 2015, 56, 1481–1489. [Google Scholar] [CrossRef] [Green Version]
- Scuffi, D.; Nietzel, T.; Di Fino, L.M.; Meyer, A.J.; Lamattina, L.; Schwarzländer, M.; Laxalt, A.M.; García-Mata, C. Hydrogen sulfide increases production of NADPH oxidase-dependent hydrogen peroxide and phospholipase D-derived phosphatidic acid in guard cell signaling. Plant Physiol. 2018, 176, 2532–2542. [Google Scholar] [CrossRef] [Green Version]
- Du, X.; Jin, Z.; Zhang, L.; Liu, X.; Yang, G.; Pei, Y. H2S is involved in ABA-mediated stomatal movement through MPK4 to alleviate drought stress in Arabidopsis thaliana. Plant Soil 2019, 435, 295–307. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, W.; Niu, J.; Ren, Y.; Zhang, F. Hydrogen sulfide may function downstream of hydrogen peroxide in salt stress-induced stomatal closure in Vicia faba. Func. Plant Biol. 2018, 46, 136–145. [Google Scholar] [CrossRef]
- Leshem, Y.Y.; Wills, R.B.H. Harnessing senescence delaying gases nitric oxide and nitrous oxide: A novel approach to post-harvest control of fresh horticultural produce. Biol. Plant. 1998, 41, 1–10. [Google Scholar] [CrossRef]
- Hung, K.T.; Kao, C.H. Nitric oxide counteracts the senescence of rice leaves induced by abscisic acid. J. Plant Physiol. 2003, 160, 871–879. [Google Scholar] [CrossRef] [Green Version]
- Corpas, F.J.; Barroso, J.B.; Carreras, A.; Quirós, M.; León, A.M.; Romero-Puertas, M.C.; Esteban, F.J.; Valderrama, R.; Palma, J.M.; Sandalio, L.M.; et al. Cellular and subcellular localization of endogenous nitric oxide in young and senescent pea plants. Plant Physiol. 2004, 136, 2722–2733. [Google Scholar] [CrossRef] [Green Version]
- Ji, Y.; Liu, J.; Xing, D. Low concentrations of salicylic acid delay methyl jasmonate-induced leaf senescence by up-regulating nitric oxide synthase activity. J. Exp. Bot. 2016, 67, 5233–5245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, Z.; Sun, L.; Yang, G.; Pei, Y. Hydrogen sulfide regulates energy production to delay leaf senescence induced by drought stress in Arabidopsis. Front. Plant Sci. 2018, 9, 1722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, K.; Peng, X.; Yao, G.; Zhou, Z.; Yang, F.; Li, W.; Zhao, Y.; Li, Y.; Han, Z.; Chen, X.; et al. Roles of a cysteine desulfhydrase LCD1 in regulating leaf senescence in tomato. Int. J. Mol. Sci. 2021, 22, 13078. [Google Scholar] [CrossRef]
- Liu, M.; Pirrello, J.; Chervin, C.; Roustan, J.P.; Bouzayen, M. Ethylene control of fruit ripening: Revisiting the complex network of transcriptional regulation. Plant Physiol. 2015, 169, 2380–2390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz-Vargas, M.A.; González-Gordo, S.; Cañas, A.; López-Jaramillo, J.; Palma, J.M.; Corpas, F.J. Endogenous hydrogen sulfide (H2S) is up-regulated during sweet pepper (Capsicum annuum L.) fruit ripening. In vitro analysis shows that NADP-dependent isocitrate dehydrogenase (ICDH) activity is inhibited by H2S and NO. Nitric Oxide 2018, 81, 36–45. [Google Scholar] [CrossRef]
- Muñoz-Vargas, M.A.; González-Gordo, S.; Palma, J.M.; Corpas, F.J. Inhibition of NADP-malic enzyme activity by H2S and NO in sweet pepper (Capsicum annuum L.) fruits. Physiol. Plant. 2020, 168, 278–288. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Du, H.; Wang, W.; Zhang, W.; Shen, Y.; Wan, C.; Chen, J. Synergistic effect of nitric oxide with hydrogen sulfide on inhibition of ripening and softening of peach fruits during storage. Sci. Hort. 2019, 256, 108591. [Google Scholar] [CrossRef]
- Huang, D.; Wang, Y.; Zhang, D.; Dong, Y.; Meng, Q.; Zhu, S.; Zhang, L. Strigolactone maintains strawberry quality by regulating phenylpropanoid, NO, and H2S metabolism during storage. Postharvest Biol. Technol. 2021, 178, 111546. [Google Scholar] [CrossRef]
- Mock, H.P.; Dietz, K.J. Redox proteomics for the assessment of redox-related post-translational regulation in plants. Biochim. Biophys. Acta—Proteins Proteom. 2016, 1864, 967–973. [Google Scholar] [CrossRef]
- Chen, J.; Shang, Y.T.; Wang, W.H.; Chen, X.Y.; He, E.M.; Zheng, H.L.; Shangguan, Z. Hydrogen sulfide-mediated polyamines and sugar changes are involved in hydrogen sulfide-induced drought tolerance in Spinacia oleracea seedlings. Front. Plant Sci. 2016, 7, 1173. [Google Scholar] [CrossRef] [Green Version]
- Du, X.; Jin, Z.; Liu, Z.; Liu, D.; Zhang, L.; Ma, X.; Yang, G.; Liu, S.; Guo, Y.; Pei, Y. H2S persulfidated and increased kinase activity of MPK4 to response cold stress in Arabidopsis. Front. Mol. Biosci. 2021, 8, 635470. [Google Scholar] [CrossRef]
- Li, J.; Shi, C.; Wang, X.; Liu, C.; Ding, X.; Ma, P.; Wang, X.; Jia, H. Hydrogen sulfide regulates the activity of antioxidant enzymes through persulfidation and improves the resistance of tomato seedling to copper oxide nanoparticles (CuO NPs)-induced oxidative stress. Plant Physiol. Biochem. 2020, 156, 257–266. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, J.; Zhou, M.; Zhou, H.; Cui, B.; Gotor, C.; Romero, L.C.; Fu, L.; Yang, J.; Foyer, C.H.; et al. Persulfidation-based modification of cysteine desulfhydrase and the NADPH oxidase RBOHD controls guard cell abscisic acid signaling. Plant Cell 2020, 32, 1000–1017. [Google Scholar] [CrossRef]
- Laureano-Marín, A.M.; Aroca, Á.; Pérez-Pérez, M.E.; Yruela, I.; Jurado-Flores, A.; Moreno, I.; Crespo, J.L.; Romero, L.C.; Gotor, C. Abscisic acid-triggered persulfidation of the Cys protease ATG4 mediates regulation of autophagy by sulfide. Plant Cell 2020, 32, 3902–3920. [Google Scholar] [CrossRef]
- Aroca, A.; Yruela, I.; Gotor, C.; Bassham, D.C. Persulfidation of ATG18a regulates autophagy under ER stress in Arabidopsis. Proc. Nat. Acad. Sci. USA 2021, 118, e2023604118. [Google Scholar] [CrossRef]
- González-Gordo, S.; Bautista, R.; Claros, M.G.; Cañas, A.; Palma, J.M.; Corpas, F.J. Nitric oxide-dependent regulation of sweet pepper fruit ripening. J. Exp. Bot. 2019, 70, 4557–4570. [Google Scholar] [CrossRef] [Green Version]
- Corpas, F.J.; González-Gordo, S.; Rodríguez-Ruiz, M.; Muñoz-Vargas, M.A.; Palma, J.M. Thiol-based oxidative posttranslational modifications (oxiPTMs) of plant proteins. Plant Cell Physiol. 2022, 63, 889–900. [Google Scholar] [CrossRef]
- Lindermayr, C.; Saalbach, G.; Durner, J. Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 2005, 137, 921–930. [Google Scholar] [CrossRef]
- Fares, A.; Rossignol, M.; Peltier, J.B. Proteomics investigation of endogenous S-nitrosylation in Arabidopsis. Biochem. Biophys. Res. Commun. 2011, 416, 331–336. [Google Scholar] [CrossRef]
- Puyaubert, J.; Fares, A.; Rézé, N.; Peltier, J.B.; Baudouin, E. Identification of endogenously S-nitrosylated proteins in Arabidopsis plantlets: Effect of cold stress on cysteine nitrosylation level. Plant Sci. 2014, 215, 150–156. [Google Scholar] [CrossRef]
- Hu, J.; Huang, X.; Chen, L.; Sun, X.; Lu, C.; Zhang, L.; Wang, Y.; Zuo, J. Site-specific nitrosoproteomic identification of endogenously S-nitrosylated proteins in Arabidopsis. Plant Physiol. 2015, 167, 1731–1746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolbert, Z.; Molnár, Á.; Oláh, D.; Feigl, G.; Horváth, E.; Erdei, L.; Ördög, A.; Rudolf, E.; Barth, T.; Lindermayr, C. S-Nitrosothiol signaling is involved in regulating hydrogen peroxide metabolism of zinc-stressed Arabidopsis. Plant Cell Physiol. 2019, 60, 2449–2463. [Google Scholar] [CrossRef]
- Rodríguez-Ruiz, M.; González-Gordo, S.; Cañas, A.; Campos, M.J.; Paradela, A.; Corpas, F.J.; Palma, J.M. Sweet pepper (Capsicum annuum L.) fruits contain an atypical peroxisomal catalase that is modulated by reactive oxygen and nitrogen species. Antioxidants 2019, 8, 374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, L.S.; Alves, M.Q.; Seabra, A.R.; Carvalho, H.G. Characterization of plant glutamine synthetase S-nitrosation. Nitric Oxide 2019, 88, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Costa-Broseta, Á.; Castillo, M.; León, J. Post-translational modifications of nitrate reductases autoregulates nitric oxide biosynthesis in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 549. [Google Scholar] [CrossRef] [PubMed]
- Jedelská, T.; Luhová, L.; Petřivalský, M. Nitric oxide signalling in plant interactions with pathogenic fungi and oomycetes. J. Exp. Bot. 2021, 72, 848–863. [Google Scholar] [CrossRef]
- González-Gordo, S.; Rodríguez-Ruiz, M.; Paradela, A.; Ramos-Fernández, A.; Corpas, F.J.; Palma, J.M. Mitochondrial protein expression during sweet pepper (Capsicum annuum L.) fruit ripening: iTRAQ-based proteomic analysis and role of cytochrome c oxidase. J. Plant Physiol. 2022, 153734. [Google Scholar] [CrossRef]
- Lounifi, I.; Arc, E.; Molassiotis, A.; Job, D.; Rajjou, L.; Tanou, G. Interplay between protein carbonylation and nitrosylation in plants. Proteomics 2013, 13, 568–578. [Google Scholar] [CrossRef]
- Ciacka, K.; Tymiński, M.; Gniazdowska, A.; Krasuska, U. Carbonylation of proteins—An element of plant ageing. Planta 2020, 252, 12. [Google Scholar] [CrossRef]
- Tola, A.J.; Jaballi, A.; Missihoun, T.D. Protein carbonylation: Emerging roles in plant redox biology and future prospects. Plants 2021, 10, 1451. [Google Scholar] [CrossRef]
- Moller, I.M.; Havelund, J.F.; Rogowska-Wrzesinska, A. Protein carbonylation in plants. In Protein Carbonylation: Principles, Analysis, and Biological Implications; Ros, J., Ed.; Wiley: Hoboken, NJ, USA, 2017; pp. 321–339. [Google Scholar] [CrossRef]
- Matamoros, M.A.; Kim, A.; Peñuelas, M.; Ihling, C.; Griesser, E.; Hoffmann, R.; Fedorova, M.; Frolov, A.; Becana, M. Protein carbonylation and glycation in legume nodules. Plant Physiol. 2018, 177, 1510–1528. [Google Scholar] [CrossRef] [Green Version]
- Satour, P.; Youssef, C.; Châtelain, E.; Vu, B.L.; Teulat, B.; Job, C.; Job, D.; Montrichard, F. Patterns of protein carbonylation during Medicago truncatula seed maturation. Plant Cell Environ. 2018, 41, 2183–2194. [Google Scholar] [CrossRef]
- Álvarez-Robles, M.J.; Bernal, M.P.; Sánchez-Guerrero, A.; Sevilla, F.; Clemente, R. Major As species, lipid peroxidation and protein carbonylation in rice plants exposed to increasing As (V) concentrations. Heliyon 2020, 6, e04703. [Google Scholar] [CrossRef]
- Li, B.B.; Zhang, S.B.; Lv, Y.Y.; Wei, S.; Hu, Y.S. Reactive oxygen species-induced protein carbonylation promotes deterioration of physiological activity of wheat seeds. PLoS ONE 2022, 17, e0263553. [Google Scholar] [CrossRef]
- Jaballi, A.; Missihoun, T.D. The phytohormone abscisic acid modulates protein carbonylation in Arabidopsis thaliana. Physiol. Plant. 2022, 174, e13658. [Google Scholar] [CrossRef]
- Hemsley, P.A. Assaying protein S-acylation in plants. In G Protein-Coupled Receptor Signaling in Plants; Humana Press: Totowa, NJ, USA, 2013; pp. 141–146. [Google Scholar] [CrossRef]
- Li, Y.; Qi, B. Progress toward understanding protein S-acylation: Prospective in plants. Front. Plant Sci. 2017, 8, 346. [Google Scholar] [CrossRef]
- Srivastava, V.; Weber, J.R.; Malm, E.; Fouke, B.W.; Bulone, V. Proteomic analysis of a poplar cell suspension culture suggests a major role of protein S-acylation in diverse cellular processes. Front. Plant Sci. 2016, 477. [Google Scholar] [CrossRef] [Green Version]
- Zheng, L.; Liu, P.; Liu, Q.; Wang, T.; Dong, J. Dynamic protein S-acylation in plants. Int. J. Mol. Sci. 2019, 20, 560. [Google Scholar] [CrossRef] [Green Version]
- Kumar, M.; Carr, P.; Turner, S.R. An atlas of Arabidopsis protein S-acylation reveals its widespread role in plant cell organization and function. Nat. Plants 2022, 8, 670–681. [Google Scholar] [CrossRef]
- Hurst, C.H.; Wright, K.M.; Turnbull, D.; Leslie, K.; Jones, S.; Hemsley, P.A. Juxta-membrane S-acylation of plant receptor-like kinases is likely fortuitous and does not necessarily impact upon function. Sci. Rep. 2019, 9, 12818. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Ahsan, N.; Thelen, J.J.; Stacey, G. S-Acylation of plant immune receptors mediates immune signaling in plasma membrane nanodomains. bioRxiv 2019, 720482. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Li, M.; Li, Y.; Chen, Z.; Zhuge, C.; Ouyang, Y.; Zhao, Y.; Lin, Y.; Xie, Q.; Yang, C.; et al. An ABHD17-like hydrolase screening system to identify de-S-acylation enzymes of protein substrates in plant cells. Plant Cell 2021, 33, 3235–3249. [Google Scholar] [CrossRef] [PubMed]
- Hurst, C.H.; Turnbull, D.; Gronnier, J.; Myles, S.; Pflughaupt, R.L.; Kopischke, M.; Davies, P.; Jones, S.; Robatzek, S.; Zipfel, C.; et al. S-acylation is a positive regulator of FLS2-mediated plant immunity. bioRxiv 2021. [Google Scholar] [CrossRef]
- Chen, D.; Hao, F.; Mu, H.; Ahsan, N.; Thelen, J.J.; Stacey, G. S-acylation of P2K1 mediates extracellular ATP-induced immune signaling in Arabidopsis. Nat. Commun. 2021, 12, 2750. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Jacobsen, S.E. Epigenetic modifications in plants: An evolutionary perspective. Curr. Opin. Plant Biol. 2011, 14, 179–186. [Google Scholar] [CrossRef] [Green Version]
- Baulcombe, D.C.; Dean, C. Epigenetic regulation in plant responses to the environment. Cold Spring Harb. Perspect. Biol. 2014, 6, 019471. [Google Scholar] [CrossRef]
- Liang, Z.; Riaz, A.; Chachar, S.; Ding, Y.; Du, H.; Gu, X. Epigenetic modifications of mRNA and DNA in plants. Mol. Plant 2020, 13, 14–30. [Google Scholar] [CrossRef]
- Turkan, I. ROS and RNS: Key signalling molecules in plants. J. Exp. Bot. 2018, 69, 3313. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishnan, M.; Papolu, P.K.; Satish, L.; Vinod, K.K.; Wei, Q.; Sharma, A.; Emamverdian, A.; Zou, L.H.; Zhou, M. Redox status of the plant cell determines epigenetic modifications under abiotic stress conditions and during developmental processes. J. Adv. Res. 2022, 42, 99–116. [Google Scholar] [CrossRef]
- Shen, Y.; Issakidis-Bourguet, E.; Zhou, D.X. Perspectives on the interactions between metabolism, redox, and epigenetics in plants. J. Exp. Bot. 2016, 67, 5291–5300. [Google Scholar] [CrossRef]
- Huang, L.; Sun, Q.; Qin, F.; Li, C.; Zhao, Y.; Zhou, D.X. Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol. 2007, 144, 1508–1519. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Xu, W.; Wang, C.; Yi, X.; Zhang, W.; Su, Z. Differential deposition of H2A. Z in combination with histone modifications within related genes in Oryza sativa callus and seedling. Plant J. 2017, 89, 264–277. [Google Scholar] [CrossRef] [Green Version]
- Yagci, S.; Yildirim, E.; Yildirim, N.; Shams, M.; Agar, G. Nitric oxide alleviates the effects of copper-induced DNA methylation, genomic instability, LTR retrotransposon polymorphism and enzyme activity in lettuce. Plant Physiol. Rep. 2019, 24, 289–295. [Google Scholar] [CrossRef]
- Zheng, S.; Li, J.; Ma, L.; Wang, H.; Zhou, H.; Ni, E.; Jiang, D.; Liu, Z.; Zhuang, C. OsAGO2 controls ROS production and the initiation of tapetal PCD by epigenetically regulating OsHXK1 expression in rice anthers. Proc. Nat. Acad. Sci. USA. 2019, 116, 7549–7558. [Google Scholar] [CrossRef] [Green Version]
- Guo, D.L.; Li, Q.; Zhao, H.L.; Wang, Z.G.; Zhang, G.H.; Yu, Y.H. The variation of berry development and DNA methylation after treatment with 5-azaC on ‘Kyoho’grape. Sci. Hort. 2019, 246, 265–271. [Google Scholar] [CrossRef]
- Pérez-Pizá, M.C.; Ibañez, V.N.; Varela, A.; Cejas, E.; Ferreyra, M.; Chamorro-Garces, J.C.; Zilli, C.; Vallecorsa, P.; Fina, B.; Prevosto, L.; et al. Non-thermal plasmas affect plant growth and DNA methylation patterns in Glycine max. J. Plant Growth Regul. 2021, 41, 2732–2742. [Google Scholar] [CrossRef]
- Villagómez-Aranda, A.L.; García-Ortega, L.F.; Torres-Pacheco, I.; Guevara-González, R.G. Whole-Genome D.N.A. methylation analysis in hydrogen peroxide overproducing transgenic tobacco resistant to biotic and abiotic stresses. Plants 2021, 10, 178. [Google Scholar] [CrossRef]
- Marvasi, M. Potential use and perspectives of nitric oxide donors in agriculture. J. Sci. Food Agric. 2017, 97, 1065–1072. [Google Scholar] [CrossRef]
- Seabra, A.B.; Rai, M.; Durán, N. Nano carriers for nitric oxide delivery and its potential applications in plant physiological process: A mini review. J. Plant Biochem. Biotechnol. 2014, 23, 1–10. [Google Scholar] [CrossRef]
- Bethke, P.C.; Libourel, I.G.; Jones, R.L. Nitric oxide reduces seed dormancy in Arabidopsis. J. Exp. Bot. 2006, 57, 517–526. [Google Scholar] [CrossRef]
- Arc, E.; Galland, M.; Godin, B.; Cueff, G.; Rajjou, L. Nitric oxide implication in the control of seed dormancy and germination. Front. Plant Sci. 2013, 4, 346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagel, M.; Alqudah, A.M.; Bailly, M.; Rajjou, L.; Pistrick, S.; Matzig, G.; Börner, A.; Kranner, I. Novel loci and a role for nitric oxide for seed dormancy and preharvest sprouting in barley. Plant Cell Environ. 2019, 42, 1318–1327. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Zhang, Y.; Liu, L.; Liu, X.; Li, B.; Jin, C.; Lin, X. Molecular functions of nitric oxide and its potential applications in horticultural crops. Hort. Res. 2021, 8, 71. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.H.; Wang, Y.; Ye, X.Y.; Li, Z.G. Signaling molecule hydrogen sulfide improves seed germination and seedling growth of maize (Zea mays L.) under high temperature by inducing antioxidant system and osmolyte biosynthesis. Front. Plant Sci. 2018, 9, 1288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caverzan, A.; Ebone, L.A.; Chiomento, J.L.T.; Chavarria, G. Physiological role of hydrogen sulfide in seed germination and seedling development under stress conditions. Arch. Agron. Soil Sci. 2021, 1530–1542. [Google Scholar] [CrossRef]
- Manjunatha, G.; Lokesh, V.; Neelwarne, B. Nitric oxide in fruit ripening: Trends and opportunities. Biotechnol. Adv. 2010, 28, 489–499. [Google Scholar] [CrossRef]
- Palma, J.M.; Freschi, L.; Rodríguez-Ruiz, M.; González-Gordo, S.; Corpas, F.J. Nitric oxide in the physiology and quality of fleshy fruits. J. Exp. Bot. 2019, 70, 4405–4417. [Google Scholar] [CrossRef]
- Jayarajan, S.; Sharma, R.R. Impact of nitric oxide on shelf life and quality of nectarine (Prunus persica var. nucipersica). Acta Physiol. Plant. 2018, 40, 207. [Google Scholar] [CrossRef]
- Sahu, S.K.; Barman, K.; Singh, A.K. Nitric oxide application for post-harvest quality retention of guava fruits. Acta Physiol. Plant. 2020, 42, 156. [Google Scholar] [CrossRef]
- Yao, G.F.; Li, C.; Sun, K.K.; Tang, J.; Huang, Z.Q.; Yang, F.; Huang, G.G.; Hu, L.Y.; Jin, P.; Hu, K.D.; et al. Hydrogen sulfide maintained the good appearance and nutrition in post-harvest tomato fruits by antagonizing the effect of ethylene. Front. Plant Sci. 2020, 11, 584. [Google Scholar] [CrossRef]
- Molinett, S.A.; Alfaro, J.F.; Sáez, F.A.; Elgueta, S.; Moya-León, M.A.; Figueroa, C.R. Post-harvest treatment of hydrogen sulfide delays the softening of chilean strawberry fruit by downregulating the expression of key genes involved in pectin catabolism. Int. J. Mol. Sci. 2021, 22, 10008. [Google Scholar] [CrossRef]
- Siddiqui, M.H.; Khan, M.N.; Mukherjee, S.; Basahi, R.A.; Alamri, S.; Al-Amri, A.A.; Alsubaie, Q.D.; Ali, H.M.; Al-Munqedhi, B.M.A.; Almohisen, I.A.A. Exogenous melatonin-mediated regulation of K+/Na+ transport, H+-ATPase activity and enzymatic antioxidative defence operate through endogenous hydrogen sulphide signalling in NaCl-stressed tomato seedling roots. Plant Biol. 2021, 23, 797–805. [Google Scholar] [CrossRef]
- Lata, D.; Homa, F.; Nayyer, M.A.; Kumar, A.; Aftab, M.A.; Siddiqui, M.W. Effect of post-harvest hydrogen sulphide on lignification and biochemical markers of pointed gourd. Plant Biol. 2022. [Google Scholar] [CrossRef]
- Fang, L.; Ju, W.; Yang, C.; Jin, X.; Liu, D.; Li, M.; Yu, J.; Zhao, W.; Zhang, C. Exogenous application of signaling molecules to enhance the resistance of legume-rhizobium symbiosis in Pb/Cd-contaminated soils. Environ. Pollut. 2020, 265, 114744. [Google Scholar] [CrossRef]
- Baudouin, E.; Pieuchot, L.; Engler, G.; Pauly, N.; Puppo, A. Nitric oxide is formed in Medicago truncatula-Sinorhizobium meliloti functional nodules. Mol. Plant Microbe Interact. 2006, 19, 970–975. [Google Scholar] [CrossRef] [Green Version]
- Nagata, M.; Murakami, E.I.; Shimoda, Y.; Shimoda-Sasakura, F.; Kucho, K.I.; Suzuki, A.; Abe, M.; Higashi, S.; Uchiumi, T. Expression of a class 1 hemoglobin gene and production of nitric oxide in response to symbiotic and pathogenic bacteria in Lotus japonicus. Mol. Plant Microbe Interact. 2008, 21, 1175–1183. [Google Scholar] [CrossRef]
- Del Giudice, J.; Cam, Y.; Damiani, I.; Fung-Chat, F.; Meilhoc, E.; Bruand, C.; Brouquisse, R.; Puppo, A.; Boscari, A. Nitric oxide is required for an optimal establishment of the Medicago truncatula–Sinorhizobium meliloti symbiosis. New Phytol. 2011, 191, 405–417. [Google Scholar] [CrossRef] [Green Version]
- Fukudome, M.; Shimada, H.; Uchi, N.; Osuki, K.I.; Ishizaki, H.; Murakami, E.I.; Kawaguchi, M.; Uchiumi, T. Reactive sulfur species interact with other signal molecules in root nodule symbiosis in Lotus japonicus. Antioxidants 2020, 9, 145. [Google Scholar] [CrossRef] [Green Version]
- Arthikala, M.K.; Montiel, J.; Sánchez-López, R.; Nava, N.; Cárdenas, L.; Quinto, C. Respiratory burst oxidase homolog gene a is crucial for rhizobium infection and nodule maturation and function in common bean. Front. Plant Sci. 2017, 8, 2003. [Google Scholar] [CrossRef]



| Enzymes | Reaction Catalysed | Cellular Location | Uniprot/Gene | References | |
|---|---|---|---|---|---|
| ROS | Cytochrome C oxidases and alternative oxidases | (Mehler’s reaction) | Thylakoid membrane | A0A1P8AZ61 /AT2G43780, Q39219/AT3G22370 | [20] |
| Cytochrome C oxidases and alternative oxidases | Thylakoid membrane/mitochondria | A0A1P8AZ61 /AT2G43780, Q39219/AT3G22370 | [20] | ||
| Cytochrome C oxidases and superoxide dismutase | (Haber- Weiss reaction) | Thylakoid membrane/peroxisomes | A0A1P8AZ61 /AT2G43780, P24704/AT1G08830 | [20] | |
| Cytochrome C oxidases and alternative oxidases | (Fenton reaction) | Thylakoid membrane | A0A1P8AZ61 /AT2G43780, Q39219/AT3G22370 | [20] | |
| RNS | Nitrate reductase | Peroxisomes | P11035 /AT1G37130, | [8,21] | |
| NOS-like activity, aldehyde oxidase, sulfite oxidase, and xanthine dehydrogenase | Chloroplast, mitochondria | Q7G191/AT1G04580, Q8GUQ8 /AT4G34890 | [8,21] | ||
| Peroxidase | Chloroplast, mitochondria | Q9SMU8/AT3G49120 | [8,21] | ||
| Amidoxime reducing components | Chloroplast, mitochondria | LOC9299625 | [8,21] | ||
| RSS | L-cysteine desulfhydrase | Cytosol | Q9MIR1/At3g62130 | [11] | |
| D-cysteine desulfhydrase | Mitochondria | A1L4V7/At3g26115 | [11] | ||
| Sulfite reductase | Chloroplast | Q9LZ66/At5g04590 | [11] | ||
| Cyanoalanine synthase | Mitochondria | Q9S757/At3g61440 | [11] | ||
| Cysteine synthase 1 | cytosol | P47998/At4g14880 | [11] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rai, K.K.; Kaushik, P. Free Radicals Mediated Redox Signaling in Plant Stress Tolerance. Life 2023, 13, 204. https://doi.org/10.3390/life13010204
Rai KK, Kaushik P. Free Radicals Mediated Redox Signaling in Plant Stress Tolerance. Life. 2023; 13(1):204. https://doi.org/10.3390/life13010204
Chicago/Turabian StyleRai, Krishna Kumar, and Prashant Kaushik. 2023. "Free Radicals Mediated Redox Signaling in Plant Stress Tolerance" Life 13, no. 1: 204. https://doi.org/10.3390/life13010204
APA StyleRai, K. K., & Kaushik, P. (2023). Free Radicals Mediated Redox Signaling in Plant Stress Tolerance. Life, 13(1), 204. https://doi.org/10.3390/life13010204