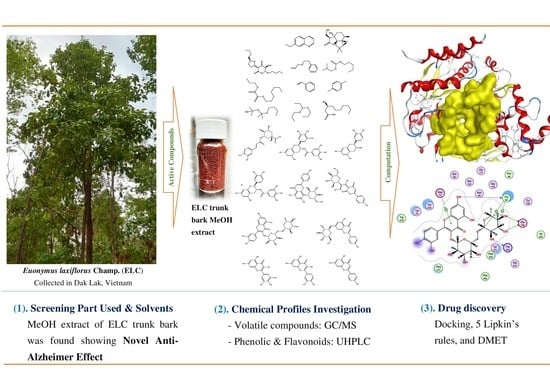
Novel Anti-Acetylcholinesterase Effect of Euonymus laxiflorus Champ. Extracts via Experimental and In Silico Studies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Chemical Methods
2.2.1. Preparation of Herbal Extracts
2.2.2. Gas Chromatography-Mass Spectrometry (GCMS) Analysis
2.2.3. UHPLC Analysis
2.3. Enzyme Inhibition Assay
2.4. Virtual Study Methods
2.4.1. Docking Simulation
- ✓
- Preparation of protein structure: The structure data of enzyme AChE were obtained from the Research Collaboratory for Structural Bioinformatics Protein Data Bank for the preparation of their three-dimensional structures through the use of MOE-2015.10. The most active sites on the enzyme were found using the site finder function of the MOE software. A virtual pH 7 was used for the preparation of enzyme structure.
- ✓
- Preparation of ligand structures: The structures of identified compounds (ligands) from the herbal extracts and commercial inhibitors were prepared using the ChemBioOffice 2018 software and further optimized using the MOE software. The following parameters were required to prepare the structure of ligands: virtual pH 7, force field MMFF94x; R-Field 1:80; cutoff, rigid water molecules, space group p1, cell size 10, 10, 10; cell shape 90, 90, 90; gradient 0.01 RMS kcal·mol−1A−2.
- ✓
- Docking ligands into enzymes and the obtained out-put data: The prepared ligands were docked into the active site of AChE using the MOE software. As major out-put for analysis, DS value, RMSD value, linkage types, compositions of amino acids, and the linkages’ distances were harvested.
2.4.2. The Lipinski Rule of Five and ADMET Analyses
3. Results and Discussion
3.1. New Records of the Potential Anti-Acetylcholinesterase Effect of Euonymus laxiflorus Champ
3.2. The Chemical Profile of the MeOH Extract of Euonymus laxiflorus Champ. Trunk Bark
3.3. Insight into the Interactions and Energy Binding Bioactive Compounds toward Targeting Enzyme—Acetylcholinesterase via Docking Study
3.4. Lipinski’s Rule of Five and ADMET-Based Pharmacokinetics and Pharmacology
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A




| Name of Protein | Surface Area (SA) Å2 | Volume (SA) Å3 |
|---|---|---|
| Electrophorus electricus AChE (1EEA) | 529.676 | 904.278 |
| Amino acids located at the active site of 1EEA: PRO229 ASN230 CYS231 PRO232 TRP233 SER 235 VAL236 SER237 GLU240 ARG244 LEU282 PRO283 PHE284 SER286 ARG289 PHE290 VAL293 ILE296 SER304 LEU305 GLU306 PRO361 HIS362 HIS398 CYS402 PRO403 HIS406 TRP524 ASN525 GLN526 LEU 528 PRO529 LEU532 ASN533 | ||

| SITE | SIZE | Residues |
|---|---|---|
| 1 | 202 | 1:(GLN69 TYR70 VAL71 ASP72 GLN74 SER81 TRP84 ASN85 PRO86 TYR116 GLY117 GLY118 GLY119 TYR121 SER122 GLY123 SER124 LEU127 TYR130 GLU199 SER200 TRP233 TRP279 LEU282 PHE284 ASP285 SER286 ILE287 PHE288 ARG289 PHE290 PHE330 PHE331 TYR334 GLY335 HIS440 GLY441 TYR442 ILE444) |
| 2 | 39 | 1:(ASN230 PRO232 GLU306 ASP397 HIS398 CYS402 PRO403 HIS406 TRP524 ASN525 PRO529) |
| 3 | 32 | 1:(LYS325 ASP326 ARG388 ASP389 ASP392 ASP393 ILE401 PHE422 GLU434 TRP435 ARG517) |
| 4 | 20 | 1:(PRO232 GLU240 ARG244 LEU282 PRO283 PHE284 ASP285 SER286 ARG289 PRO361 HIS362) |
| 5 | 20 | 1:(ARG349 PHE352 TYR375 THR376 ASP377 ASP380 ASP381 LYS386 ASN387 GLY390 LEU391) |
| 6 | 27 | 1:(CYS402 MET405 HIS406 ASN409 LYS410 GLN500 ARG515 LEU516 ARG517 VAL518 CYS521 VAL522 ASN525 GLN526) |
| 7 | 19 | 1:(ALA36 GLU37 PRO38 PRO39 MET43 ARG46 ARG47 PRO48 GLU49 LEU95 ARG149) |
| 8 | 17 | 1:(ARG468 TYR472 SER487 GLU489 SER490 GLU508 PRO509 MET510) |
| 9 | 22 | 1:(ARG47 GLY166 ASN167 LEU171 ARG174 SER212 PRO213 GLY214 ASP297 GLU299 PHE300) |
| 10 | 19 | 1:(THR412 GLY415 ASN416 GLY417 THR418 LEU494 PHE495 THR496 THR497) |
| 11 | 16 | 1:(GLU461 ALA464 LEU465 ARG468 ASN506 THR507 GLU508 PRO509) |
| 12 | 24 | 1:(PRO39 ARG44 CYS67 GLN68 SER91 GLU92 CYS94 TYR148 ARG149 VAL150 PHE153) |
| 13 | 12 | 1:(GLN68 GLN69 TYR70 TYR121 VAL150 GLY151 ALA152 PHE153 LEU274 ILE275 GLU278) |
| 14 | 18 | 1:(GLU82 MET83 ASN85 ASN87 LEU127 ASP128 VAL129) |
| 15 | 17 | 1:(GLN69 TYR70 VAL71 GLN272 ILE275 ASP276) |
| 16 | 19 | 1:(LEU450 PRO451 LEU452 VAL453 LYS454 LEU456 ASN457 TYR458 THR459 ALA460 GLU463) |
| 17 | 23 | 1:(ASP128 VAL129 ASN131 LYS133 TYR134 LEU450 VAL453 GLU455 LEU456) |
| 18 | 20 | 1:(TYR375 THR376 ASP377 LYS386 ASP389 GLY390 ASP393 ARG517 MET520) |
| 19 | 12 | 1:(LEU31 GLY32 TRP58 ASN59 ALA60 SER61 THR62 TYR63 PRO64) |
| 20 | 21 | 1:(GLN318 ASN416 GLY417 TYR419 THR479 GLY480 ASN481 LEU494) |
| 21 | 41 | 1:(MET83 VAL129 ASN429 LEU430 VAL431 TYR442 GLU445 LEU450 LEU456 TYR458) |
| 22 | 7 | 1:(LYS11 SER12 LYS51 TRP179 ASP182 ASN183) |
| 23 | 12 | 1:(GLN374 GLN519 MET520 VAL522 PHE523 PHE527) |
| 24 | 17 | 1:(ASN42 GLU163 GLU260 ILE263 HIS264 ARG267) |
| 25 | 16 | 1:(ARG47 PRO48 LEU171 ARG174 MET175 GLN178 LEU218) |
| ID Compd | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | Unit |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Property | ||||||||||||
| Absorption | ||||||||||||
| Water solubility | −2.327 | −2.051 | −1.454 | −2.522 | −2.739 | −2.3 | −2.3 | −2.633 | −2.777 | −2.445 | −2.449 | (1) |
| Caco2 permeability | 1.398 | 1.653 | 1.726 | 1.547 | 1.544 | 1.45 | 1.755 | 1.563 | 0.336 | 1.619 | −0.84 | (2) |
| Intestinal absorption (human) | 93.346 | 95.839 | 93.25 | 95.713 | 95.902 | 95.192 | 93.991 | 97.018 | 78.829 | 97.318 | 36.377 | (3) |
| Skin Permeability | −2.713 | −2.144 | −3.472 | −1.236 | −1.104 | −2.082 | −2.018 | −1.505 | −3.257 | −1.642 | −2.735 | (4) |
| P-glycoprotein substrate | No | No | No | No | No | No | No | No | No | Yes | Yes | (5) |
| P-glycoprotein I inhibitor | No | No | No | No | No | No | No | No | No | No | No | (5) |
| P-glycoprotein II inhibitor | No | No | No | No | No | No | No | No | No | No | No | (5) |
| Distribution | ||||||||||||
| VDss (human) | −0.799 | 0.054 | −0.075 | 0.325 | 0.403 | −0.034 | −0.046 | 0.414 | 0.206 | 0.184 | 0.581 | (6) |
| Fraction unbound (human) | 0.454 | 0.55 | 0.603 | 0.362 | 0.331 | 0.454 | 0.489 | 0.284 | 0.622 | 0.27 | 0.658 | (6) |
| BBB permeability | 0.252 | 0.63 | 0.005 | 0.409 | 0.459 | 0.555 | 0.525 | 0.688 | −0.008 | 0.396 | −1.407 | (7) |
| CNS permeability | −2.92 | −2.69 | −3.083 | −1.677 | −1.577 | −2.849 | −2.331 | −1.884 | −3.367 | −1.92 | −3.856 | (8) |
| Metabolism | ||||||||||||
| CYP2D6 substrate | No | No | No | No | No | No | No | No | No | No | No | (5) |
| CYP3A4 substrate | No | No | No | No | No | No | No | No | No | No | No | (5) |
| CYP1A2 inhibitor | No | No | No | No | Yes | No | No | Yes | No | Yes | No | (5) |
| CYP2C19 inhibitor | No | No | No | No | No | No | No | No | No | Yes | No | (5) |
| CYP2C9 inhibitor | No | No | No | No | No | No | No | No | No | No | No | (5) |
| CYP2D6 inhibitor | No | No | No | No | No | No | No | No | No | No | No | (5) |
| CYP3A4 inhibitor | No | No | No | No | No | No | No | No | No | No | No | (5) |
| Excretion | ||||||||||||
| Total Clearance | 1.598 | 1.639 | 1.35 | 0.254 | 0.265 | 1.774 | 0.535 | 0.35 | 0.803 | 0.296 | 0.307 | (9) |
| Renal OCT2 substrate | No | No | No | No | No | No | No | No | No | No | No | (5) |
| Toxicity | ||||||||||||
| AMES toxicity | No | No | No | No | No | No | No | No | Yes | Yes | No | (5) |
| Max. tolerated dose (human) | 0.084 | 0.796 | 0.848 | 0.921 | 0.943 | 0.771 | 0.805 | 0.999 | 0.529 | 0.331 | −0.134 | (10) |
| hERG I inhibitor | No | No | No | No | No | No | No | No | No | No | No | (5) |
| hERG II inhibitor | No | No | No | No | No | No | No | No | No | No | No | (5) |
| Oral Rat Acute Toxicity (LD50) | 1.342 | 2.004 | 1.949 | 1.841 | 1.83 | 1.947 | 2.189 | 1.828 | 3.097 | 2.023 | 1.973 | (11) |
| Oral Rat Chronic Toxicity | 2.552 | 2.269 | 2.105 | 2.168 | 2.225 | 2.087 | 2.043 | 1.946 | 1.901 | 2.11 | 2.982 | (12) |
| Hepatotoxicity | No | No | No | No | No | No | No | No | No | No | No | (5) |
| Skin Sensitization | No | Yes | Yes | No | No | Yes | Yes | Yes | No | No | No | (5) |
| T.Pyriformis toxicity | 0.38 | 0.596 | 0.181 | −0.022 | −0.018 | 0.163 | 0.833 | 0.96 | 0.31 | 0.453 | 0.285 | (13) |
| Minnow toxicity | 0.711 | 1.336 | 1.956 | 1.31 | 1.136 | 0.477 | 0.854 | 0.67 | 2.574 | 0.643 | 5.741 | (14) |
| ID Compd | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | Unit |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Property | ||||||||||||
| Absorption | ||||||||||||
| Water solubility | −2.894 | −3.117 | −2.911 | −2.845 | −2.812 | −2.892 | −2.559 | −2.915 | −2.925 | −3.329 | −6.715 | (1) |
| Caco2 permeability | −1.521 | −0.283 | −1.264 | −0.956 | −0.618 | −0.949 | 0.33 | 0.095 | −0.229 | 1.007 | 1.212 | (2) |
| Intestinal absorption (human) | 47.395 | 68.829 | 62.096 | 46.695 | 64.729 | 23.446 | 37.609 | 65.93 | 77.207 | 93.25 | 94.642 | (3) |
| Skin Permeability | −2.735 | −2.735 | −2.735 | −2.735 | −2.735 | −2.735 | −2.735 | −2.735 | −2.735 | −2.735 | −2.781 | (4) |
| P-glycoprotein substrate | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | (5) |
| P-glycoprotein I inhibitor | No | No | No | No | No | No | No | No | No | No | Yes | (5) |
| P-glycoprotein II inhibitor | Yes | No | Yes | No | No | No | No | No | No | No | Yes | (5) |
| Distribution | ||||||||||||
| VDss (human) | 0.806 | 1.027 | 0.664 | 1.071 | 1.239 | 1.663 | 0.342 | 1.317 | 1.559 | 0.822 | 0.179 | (6) |
| Fraction unbound (human) | 0.215 | 0.235 | 0.158 | 0.242 | 0.21 | 0.187 | 0.218 | 0.238 | 0.206 | 0.147 | 0 | (6) |
| BBB permeability | −2.184 | −1.054 | −1.847 | −1.449 | −1.375 | −1.899 | −1.391 | −1.493 | −1.098 | −0.734 | 0.764 | (7) |
| CNS permeability | −3.96 | −3.298 | −3.743 | −3.834 | −3.754 | −5.178 | −3.746 | −3.709 | −3.065 | −2.061 | −1.652 | (8) |
| Metabolism | ||||||||||||
| CYP2D6 substrate | No | No | No | No | No | No | No | No | No | No | No | (5) |
| CYP3A4 substrate | No | No | No | No | No | No | No | No | No | No | Yes | (5) |
| CYP1A2 inhibitor | No | No | No | No | No | No | No | Yes | Yes | Yes | No | (5) |
| CYP2C19 inhibitor | No | No | No | No | No | No | No | No | No | Yes | No | (5) |
| CYP2C9 inhibitor | No | No | No | No | No | No | No | No | No | No | No | (5) |
| CYP2D6 inhibitor | No | No | No | No | No | No | No | No | No | No | No | (5) |
| CYP3A4 inhibitor | Yes | No | No | No | No | No | No | No | No | No | No | (5) |
| Excretion | ||||||||||||
| Total Clearance | 0.292 | 0.183 | −0.169 | 0.444 | 0.442 | −0.369 | 0.547 | 0.422 | 0.407 | 0.566 | 0.619 | (9) |
| Renal OCT2 substrate | No | No | No | No | No | No | No | No | No | No | No | (5) |
| Toxicity | ||||||||||||
| AMES toxicity | No | No | No | No | No | No | No | No | No | No | No | (5) |
| Max. tolerated dose (human) | 0.441 | 0.438 | 0.449 | 0.577 | 0.649 | 0.452 | 0.515 | 0.51 | 0.499 | 0.328 | −0.653 | (10) |
| hERG I inhibitor | No | No | No | No | No | No | No | No | No | No | No | (5) |
| hERG II inhibitor | Yes | No | Yes | No | No | Yes | No | No | No | No | Yes | (5) |
| Oral Rat Acute Toxicity (LD50) | 2.522 | 2.428 | 2.558 | 2.595 | 2.558 | 2.491 | 2.595 | 2.497 | 2.471 | 2.45 | 2.553 | (11) |
| Oral Rat Chronic Toxicity | 3.065 | 2.5 | 2.777 | 4.635 | 5.37 | 3.673 | 4.359 | 2.718 | 2.612 | 2.298 | 0.89 | (12) |
| Hepatotoxicity | No | No | No | No | No | No | No | No | No | No | No | (5) |
| Skin Sensitization | No | No | No | No | No | No | No | No | No | No | No | (5) |
| T.Pyriformis toxicity | 0.285 | 0.347 | 0.285 | 0.285 | 0.285 | 0.285 | 0.285 | 0.286 | 0.288 | 0.38 | 0.432 | (13) |
| Minnow toxicity | 7.713 | 3.585 | 6.146 | 4.897 | 5.18 | 7.677 | 5.507 | 5.023 | 3.721 | 2.432 | −1.711 | (14) |
References
- Patterson, C. World Alzheimer Report 2018; Alzheimer’s Disease International: London, UK, 2018. [Google Scholar]
- Breijyeh, Z.; Karaman, R. Comprehensive review on Alzheimer’s disease: Causes and treatment. Molecules 2020, 25, 5789. [Google Scholar] [CrossRef]
- Majid, H.; Silva, F.V.M. Inhibition of enzymes important for Alzheimer’s disease by antioxidant extracts prepared from 15 New Zealand medicinal trees and bushes. J. R. Soc. N. Z. 2020, 50, 538–551. [Google Scholar] [CrossRef]
- Chen, B.W.; Li, W.X.; Wang, G.H.; Li, G.H.; Liu, J.Q.; Zheng, J.J.; Wang, Q.; Li, H.J.; Dai, S.X.; Huang, J.F. A strategy to find novel candidate anti-Alzheimer’s disease drugs by constructing interaction networks between drug targets and natural compounds in medical plants. PeerJ 2018, 6, e4756. [Google Scholar] [CrossRef] [PubMed]
- Singhal, A.K.; Naithani, V.; Bangar, O.P. Medicinal plants with a potential to treat Alzheimer and associated symptoms. Int. J. Nutr. Pharmacol. Neurol. Dis. 2012, 2, 84–91. [Google Scholar] [CrossRef]
- Tuzimski, T.; Petruczynik, A. Determination of anti-Alzheimer’s disease activity of selected plant ingredients. Molecules 2022, 27, 3222. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.K.; Kumar, V.; Houghton, P.J. Screening of Indian medicinal plants for acetylcholinesterase inhibitory activity. Phytother. Res. 2007, 21, 1142–1145. [Google Scholar] [CrossRef]
- Elufioye, T.O.; Obuotor, E.M.; Sennuga, A.T.; Joseph, M.A.; Saburi, A.A. Acetylcholinesterase and butyrylcholinesterase inhibitory activity of some selected Nigerian medicinal plants. Rev. Bras. Farmacogn. 2010, 20, 472–477. [Google Scholar] [CrossRef]
- Mack, M.; Ashwell, R.N.; Jeffrey, F.F.; Johannes, V.S. Phenolic composition. antioxidant and acetylcholinesterase inhibitory activities of Sclerocarya birrea and Harpephyllum caffrum (Anacardiaceae) extracts. Food Chem. 2010, 123, 69–76. [Google Scholar]
- Llorent, M.E.; Ortega, B.P.; Zengin, G.; Mocan, A.; Simirgiotis, M.; Ceylan, R.; Uysal, S.; Aktumsek, A. Evaluation of antioxidant potential, enzyme inhibition activity and phenolic profile of Lathyrus cicera and Lathyrus digitatus: Potential sources of bioactive compounds for the food industry. Food Chem. Toxicol. 2017, 107, 609–619. [Google Scholar] [CrossRef]
- Nguyen, D.N.V.; Nguyen, T. An Overview of the Use of Plants and Animals in Traditional Medicine Systems in Viet Nam; Traffic Southeast Asia: Petaling Jaya, Malaysia, 2008. [Google Scholar]
- Nguyen, V.B.; Nguyen, Q.V.; Nguyen, A.D.; Wang, S.L. Screening and evaluation of α-glucosidase inhibitors from indigenous medicinal plants in Dak Lak Province, Vietnam. Res. Chem. Intermed. 2017, 43, 3599–3612. [Google Scholar] [CrossRef]
- Nguyen, V.B.; Wang, S.L.; Nguyen, A.D.; Vo, T.P.K.; Zhang, L.J.; Nguyen, Q.V.; Kuo, Y.H. Isolation and identification of novel α-amylase inhibitors from Euonymus laxiflorus Champ. Res. Chem. Intermed. 2018, 44, 1411–1424. [Google Scholar] [CrossRef]
- Nguyen, V.B.; Nguyen, Q.V.; Nguyen, A.D.; Wang, S.L. Porcine pancreatic α-amylase inhibitors from Euonymus laxiflorus Champ. Res. Chem. Intermed. 2017, 43, 259–269. [Google Scholar] [CrossRef]
- Lam, T.M.L.; Nguyen, T.M.T.; Nguyen, X.H.; Dang, P.H.; Nguyen, N.T.; Tran, M.H.; Nguyen, T.H.; Nguyen, N.M.; Min, B.S.; Kim, J.A.; et al. Anti-cholinesterases and memory improving effects of Vietnamese Xylia xylo-carpa. Chem. Cent. J. 2016, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.C.; Nguyen, T.T.H.; Tran, M.H.; Tran, C.L. Anti-cholinesterase activity of lycopodium alkaloids from Vietnamese Huperzia squarrosa (Forst.) Trevis. Molecules 2014, 19, 19172–19179. [Google Scholar]
- Dung, H.V.; Cuong, T.D.; Chinh, N.M.; Quyen, D.; Kim, J.A.; Byeon, J.S.; Woo, M.H.; Choi, J.S.; Min, B.S. Compounds from the aerial parts of Piper bavinum and their anti-cholinesterase activity. Arch. Pharm. Res. 2015, 38, 677–682. [Google Scholar] [CrossRef]
- Flora of China. Available online: http://www.efloras.org/florataxon.aspx?flora_id=2&taxon_id=200012808 (accessed on 24 June 2022).
- Nguyen, V.B.; Wang, S.L.; Nguyen, A.D.; Lin, Z.H.; Doan, C.T.; Tran, T.N.; Huang, H.T.; Kuo, Y.H. Bioactivity-guided purification of novel herbal antioxidant and anti-NO compounds from Euonymus laxiflorus Champ. Molecules 2019, 24, 120. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Q.V.; Nguyen, N.H.; Wang, S.L.; Nguyen, V.B.; Nguyen, A.D. Free radical scavenging and antidiabetic activities of Euonymus laxiflorus Champ. extract. Res. Chem. Intermed. 2017, 43, 5615–5624. [Google Scholar] [CrossRef]
- Nguyen, V.B.; Ton, T.Q.; Nguyen, D.N.; Nguyen, T.T.; Nguyen, T.N.; Nguyen, T.H.; Doan, C.T.; Tran, T.N.; Nguyen, M.T.; Ho, N.D.; et al. Reclamation of beneficial bioactivities of herbal antioxidant condensed tannin extracted from Euonymus laxiflorus. Res. Chem. Intermed. 2020, 46, 4751–4766. [Google Scholar] [CrossRef]
- Nguyen, V.B.; Wang, S.L.; Nguyen, T.H.; Nguyen, M.T.; Doan, C.T.; Tran, T.N.; Lin, Z.H.; Nguyen, Q.V.; Kuo, Y.H.; Nguyen, A.D. Novel potent hypoglycemic compounds from Euonymus laxiflorus Champ. and their effect on reducing plasma glucose in an ICR mouse model. Molecules 2018, 23, 1928. [Google Scholar] [CrossRef]
- Ellman, G.L.; Courtney, K.D.; Andres, V.J.; Feather, S.R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961, 7, 88–95. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Wang, S.L.; Nguyen, A.D.; Doan, M.D.; Tran, T.N.; Doan, C.T.; Nguyen, V.B. Novel α-amylase inhibitor hemi-pyocyanin produced by microbial conversion of chitinous discards. Mar. Drugs 2022, 20, 283. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.B.; Wang, S.L.; Nguyen, A.D.; Phan, T.Q.; Techato, K.; Pradit, S. Bioproduction of prodigiosin from fishery processing waste shrimp heads and evaluation of its potential bioactivities. Fishes 2021, 6, 30. [Google Scholar] [CrossRef]
- Pires, D.E.; Blundell, T.L.; Ascher, D.B. PkCSM: Predicting smallmolecule pharmacokinetic and toxicity properties using graphbased signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef] [PubMed]
- Saxena, M.; Dubey, R. Target enzyme in Alzheimer’s disease: Acetylcholinesterase inhibitors. Curr. Top. Med. Chem. 2019, 19, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Shi, J.; Zhang, X. Herbal therapy: A new pathway for the treatment of Alzheimer’s disease. Alzheimer’s Res. Therapy 2010, 2, 30. [Google Scholar] [CrossRef] [PubMed]
- Birks, J.; Grimley, E.J. Ginkgo biloba for cognitive impairment and dementia. Cochrane Database Syst. Rev. 2009, 1, CD003120. [Google Scholar] [CrossRef]
- Ihl, R.; Bachinskaya, N.; Korczyn, A.D.; Vakhapova, V.; Tribanek, M.; Hoerr, R. Efficacy and safety of a oncedaily formulation of Ginkgo biloba extract EGb 761 in dementia with neuropsychiatric features: A randomized controlled trial. Int. J. Geriatr. Psychiatry 2010, 9, 123. [Google Scholar]
- Fu, L.M.; Li, J.T. A systematic review of single Chinese herbs for Alzheimer’s disease treatment. Evid. Based Complement. Altern. Med. 2009, 2011, 640284. [Google Scholar] [CrossRef]
- Akhondzadeh, S.; Noroozian, M.; Mohammadi, M.; Ohadinia, S.; Jamshidi, A.H.; Khani, M. Salvia officinalis extract in the treatment of patients with mild to moderate Alzheimer’s disease: A double blind, randomized and placebo-controlled trial. J. Clin. Pharm. Ther. 2003, 28, 53–59. [Google Scholar] [CrossRef]
- Huijberts, G.N.; Eggink, G.; Waard, P.D.; Huisman, G.W.; Witholt, B. Pseudomonas putida KT2442 cultivated on glucose accumulates poly(3-hydroxyalkanoates) consisting of saturated and unsaturated monomers. Appl. Environ. Microbiol. 1992, 58, 536–544. [Google Scholar] [CrossRef]
- Ekundayo, O.; Ajani, F.; Seppänen-Laakso, T.; Laakso, I. Volatile constituents of Psidium Guajava l. (guava) fruits. Flavour Fragr. J. 1991, 6, 233–236. [Google Scholar] [CrossRef]
- Ronald, C.W.; Judy, C.J.; George, L.K.H. Volatile constituents of the muscadine grape (Vitis rotundifolia). Agric. Food Chem. 1982, 30, 681–684. [Google Scholar]
- Ramadan, N.S.; Wessjohann, L.A.; Mocan, A.; Vodnar, C.; Abdou, M.D.; Abdel, A.Z.; Ehrlich, A. Nutrient and sensory metabolites profiling of Averrhoa Carambola L. (Starfruit) in the context of its origin and ripening stage by GC/MS and chemometric analysis. Molecules 2020, 25, 2423. [Google Scholar] [CrossRef]
- Isaac, J.U.; Kerenhappuch, I.U.; Hauwa, A.U. Phytochemical screening, isolation, characterization of bioactive and biological activity of bungkang, (Syzygium polyanthum) root-bark essential oil. Korean J. Food Health Converg. 2020, 6, 5–21. [Google Scholar]
- Farrell, I.W.; Halsall, T.G.; Thaller, V.; Bradshaw, A.P.W.; Hanson, J.R. Structures of some new sesquiterpenoid metabolites of Marasmius alliaceus. J. Chem. Soc. Perkin Trans. 1981, 1, 1790–1793. [Google Scholar] [CrossRef]
- Awasthi, Y.C.; Mitra, C.R. Constituents of Melia indica leaves. Phytochemistry 1971, 10, 2842. [Google Scholar] [CrossRef]
- Trabelsi, N.; Oueslati, S.; Caroline, H.V.; Pierre, W.T.; Medini, F.; Mérillon, J.M.; Abdelly, C.; Ksouri, R. Phenolic contents and biological activities of Limoniastrum guyonianum fractions obtained by centrifugal partition chromatography. Ind. Crops Prod. 2013, 49, 740–746. [Google Scholar] [CrossRef]
- Margarita, H.P.; Teresa, H.; Carmen, G.C.; Estrella, I.; Rabanal, R.M. Phenolic composition of the “Mocán” Visnea mocanera L.f.). J. Agric. Food Chem. 1996, 44, 3512–3515. [Google Scholar]
- Ohashi, K.; Winarno, H.; Mukai, M.; Inoue, M.; Prana, M.S.; Simanjuntak, P.; Shibuya, H. Indonesian medicinal plants. XXV. cancer cell invasion inhibitory effects of chemical constituents in the parasitic plant Scurrula atropurpurea (Loranthaceae). Chem. Pharm. Bull. 2003, 51, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Bruce, A.B.; Gavin, C.; Bhat, U.G. Flavonoids and the relationship of Itea to the saxifragaceae. Phytochemistry 1988, 27, 2651–2653. [Google Scholar]
- Robert, H.J.; James, W.W.J. Taxonomic implications of the flavonoids of Cymophyllus fraseri (Cyperaceae). Biochem. Systermatics Ecol. 1988, 16, 521–523. [Google Scholar]
- Backheet, E.Y.; Ahmed, A.S.; Sayed, H.M. Phytochemical study of the constituents of the leaves of Ficus infectoria (Roxb.). Bull. Pharm. Sci. 2001, 24, 21–27. [Google Scholar]
- Barberán, F.A.T.; Gil, M.I.; Tomás, F.; Ferreres, F.; Arques, A. Flavonoid aglycones and glycosides from Teucrium gnaphalodes. J. Nat. Prod. 1985, 48, 859. [Google Scholar] [CrossRef]
- Fishawy, A.A.; Rawia, Z.S.A. Phytochemical and pharmacological studies of Ficus auriculata Lour. (Moraceae) cultivated in Egypt. Planta Med. 2011, 77, 184–195. [Google Scholar]
- Fischer, C.; Speth, V.; Fleig, E.S.; Neuhaus, G. Induction of zygotic polyembryos in wheat: Influence of auxin polar transport. Plant. Cell 1997, 9, 1767–1780. [Google Scholar] [CrossRef]
- Venigalla, M.; Gyengesi, E.; Münch, G. Curcumin and Apigenin—Novel and promising therapeutics against chronic neuroinflammation in Alzheimer’s disease. Neural Regen. Res. 2015, 10, 1181–1185. [Google Scholar]
- Kuo, Y.H.; Huang, H.C.; Chiou, W.F.; Shi, L.S.; Wu, T.S.; Wu, Y.C. A novel NO-production-inhibiting triterpene and cytotoxicity of known alkaloids from Euonymus laxiflorus. J. Nat. Prod. 2003, 66, 554–557. [Google Scholar] [CrossRef]
- Ding, Y.; Fang, Y.; Moreno, J.; Ramanujam, J.; Jarrell, M.; Brylinski, M. Assessing the similarity of ligand binding conformations with the contact mode score. Comput. Biol. Chem. 2016, 64, 403–413. [Google Scholar] [CrossRef]
- Chandra, B.T.M.; Rajesh, S.S.; Bhaskar, B.V.; Devi, S.; Rammohan, A.; Sivaraman, T.; Rajendra, W. Molecular docking, molecular dynamics simulation, biological evaluation and 2D QSAR analysis of flavonoids from Syzygium alternifolium as potent anti-Helicobacter pylori agents. RSC Adv. 2017, 7, 18277–18292. [Google Scholar] [CrossRef]
- Loan, H.T.P.; Bui, T.Q.; My, T.T.A.; Hai, N.T.T.; Quang, D.T.; Tat, P.V.; Hiep, D.T.; Trung, N.T.; Quy, P.T.; Nhung, N.T.A. In-Depth Investigation of a Donor-Acceptor Interaction on the Heavy-Group-14@Group-13-Diyls in transition-metal tetrylone complexes: Structure, bonding, and property. ACS Omega 2020, 5, 21271–21287. [Google Scholar] [CrossRef]
- Rosenberg, B. Electrical conductivity of proteins. Nature 1962, 193, 364–365. [Google Scholar] [CrossRef] [PubMed]
- Kharkyanen, V.N.; Petrov, E.G.; Ukrainskii, I.I. Donor-acceptor model of electron transfer through proteins. J. Theor. Biol. 1978, 73, 29–50. [Google Scholar] [CrossRef] [PubMed]
- Cordes, M.; Giese, B. Electron transfer in peptides and proteins. Chem. Soc. Rev. 2009, 38, 892–901. [Google Scholar] [CrossRef] [PubMed]
- Thao, T.T.P.; Bui, T.Q.; Hai, N.T.T.; Huynh, L.K.; Quy, P.T.; Bao, N.C.; Nhung, N.T.A. Newly synthesised oxime and lactone derivatives from Dipterocarpus alatus dipterocarpol as anti-diabetic inhibitors: Experimental bioassay-based evidence and theoretical computation-based prediction. RSC Adv. 2021, 11, 35765–35782. [Google Scholar] [CrossRef] [PubMed]
- Pagadala, N.; Syed, K.; Tuszynski, J. Software for molecular docking: A review. Biophys. Rev. 2017, 9, 91–102. [Google Scholar] [CrossRef]
- Pantsar, T.; Poso, A. Binding Affinity via Docking: Fact and Fiction. Molecules 2018, 23, 1899. [Google Scholar] [CrossRef]
- Warren, G.L.; Andrews, C.W.; Capelli, A.M.; Clarke, B.; LaLonde, J.; Lambert, M.H.; Lindvall, M.; Nevins, N.; Semus, S.F.; Senger, S.; et al. A critical assessment of docking programs and scoring functions. J. Med. Chem. 2006, 49, 5912–5931. [Google Scholar] [CrossRef]
- Marko, B.; Yunhui, G.; Joseph, B.; Hans, B.; David, H.; Clara, C.; Jérémie, M.; David, M.; Katharina, M. Prioritizing small sets of molecules for synthesis through in-silico tools: A comparison of common ranking methods. Chem. Med. Chem. 2023, 18, e202200425. [Google Scholar]
- Huang, S.; Wang, L.L.; Xue, N.N.; Li, C.; Guo, H.H.; Ren, T.K.; Zhan, Y.; Li, W.B.; Zhang, J.; Chen, X.G.; et al. Chlorogenic acid effectively treats cancers through induction of cancer cell differentiation. Theranostics 2019, 9, 6745–6763. [Google Scholar] [CrossRef]
- Kwak, T.W.; Park, S.B.; Kim, H.J.; Jeong, Y.I.; Kang, D.H. Anticancer activities of epigallocatechin-3-gallate against cholangiocarcinoma cells. Onco Targets Ther. 2017, 10, 137–144. [Google Scholar] [CrossRef]
- Zong, P.C.; John, B.S.; Ho, C.T.; Chen, K.Y. Green tea epigallocatechin gallate shows a pronounced growth inhibitory effect on cancerous cells but not on their normal counterparts. Cancer Lett. 1998, 129, 173–179. [Google Scholar]
- Babich, H.; Krupka, M.E.; Nissim, H.A.; Zuckerbraun, H.L. Differential in vitro cytotoxicity of (−)-epicatechin gallate (ECG) to cancer and normal cells from the human oral cavity. Toxicol. Vitr. 2005, 19, 231–242. [Google Scholar] [CrossRef]
- Li, Z.; Feng, C.; Dong, H.; Jin, W.; Zhang, W.; Zhan, J.; Wang, S. Health promoting activities and corresponding mechanism of (–)-epicatechin-3-gallate. Food Sci. Hum. Wellness 2022, 11, 568–578. [Google Scholar] [CrossRef]
- Hosseinimehr, S.J.; Rostamnezad, M.; Ghafarirad, V. Epicatechin enhances anti-proliferative effect of bleomycin in ovarian cancer cell. Res. Mol. Med. 2013, 1, 25–28. [Google Scholar] [CrossRef]
- Elbaz, H.A.; Lee, I.; Antwih, D.A.; Liu, J.; Hüttemann, M.; Zielske, S.P. Epicatechin stimulates mitochondrial activity and selectively sensitizes cancer cells to radiation. PLoS ONE 2014, 9, e88322. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, Q.; Liu, H. Vitexin induces apoptosis through mitochondrial pathway and PI3K/Akt/mTOR signaling in human non-small cell lung cancer A549 cells. Biol. Res. 2019, 52, 7. [Google Scholar] [CrossRef] [PubMed]
- Satari, A.; Ghasemi, S.; Habtemariam, S.; Asgharian, S.; Lorigooini, Z. Rutin: A Flavonoid as an Effective Sensitizer for Anticancer Therapy; Insights into Multifaceted Mechanisms and Applicability for Combination Therapy. Evid.-Based Complement. Altern. Med. ECAM 2021, 2021, 9913179. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Qi, M.; Li, P.; Zhan, Y.; Shao, H. Apigenin in cancer therapy: Anti-cancer effects and mechanisms of action. Cell Biosci. 2017, 7, 50. [Google Scholar] [CrossRef]
- Afroze, N.; Pramodh, S.; Hussain, A.; Waleed, M.; Vakharia, K. A review on myricetin as a potential therapeutic candidate for cancer prevention. Biotech 2020, 10, 211. [Google Scholar] [CrossRef]
- Srivastava, S.; Somasagara, R.R.; Hegde, M.; Nishana, M.; Tadi, S.K.; Srivastava, M.; Choudhary, B.; Raghavan, S.C. Quercetin, a natural flavonoid interacts with DNA, arrests cell cycle and causes tumor regression by activating mitochondrial pathway of apoptosis. Sci. Rep. 2016, 6, 24049. [Google Scholar] [CrossRef]







| No. | ELCTB Extracted by Various Solvents | IC50 (mg/mL) | Max Inhibition (%) |
|---|---|---|---|
| 1 | Methanol extract | 0.323 ± 0.021 | 97.0 ± 2.3% |
| 2 | Butanol extract | 0.781 ± 0.042 | 80.1 ± 2.4% |
| 3 | Ethyl acetate extract | 1.232 ± 0.098 | 65.2 ± 3.1% |
| 4 | n-Hexane extract | 1.530 ± 0.101 | 57.3 ± 1.5% |
| 5 | Water extract | 0.662 ± 0.032 | 79.1 ± 2.6% |
| 6 | Berberine chloride | 0.314 ± 0.032 | 98.2 ± 1.8% |
| Parts Used of Euonymus laxiflorus Champ. Extracted by MeOH | IC50 (mg/mL) | Total Content of Polyphenol (mg GAE/g Dry Extracts) | Total Content of Flavonoid (mg QE/g Dry Extracts) |
|---|---|---|---|
| Heartwood MeOH extract | 1.326 ± 0.103 | 107.2 ± 22.1 | 98.0 ± 7.8 |
| Trunk bark MeOH extract | 0.336 ± 0.029 | 567.3 ± 27.2 | 335.2 ± 10 |
| Leaves MeOH extract | 0.981 ± 0.045 | 227.2 ± 15.1 | 101.3 ± 8.9 |
| Berberine chloride | 0.327 ± 0.031 | - | - |
| ID Compd | Compds | RT (min) | % Area |
|---|---|---|---|
| 1 | 3-Hydroxydecanoic acid | 3.40 | 48.63 |
| 2 | Propane, 1,1-dipropoxy- (CAS) | 4.77 | 2.91 |
| 3 | (2-(2-butoxyisopropoxy)-2-isopropanol | 4.88 | 2.62 |
| 4 | p-Xylene | 5.16 | 11.63 |
| 5 | Styrene | 5.56 | 8.53 |
| 6 | Oxalic acid, heptyl propyl ester | 7.45 | 4.62 |
| 7 | Sulfurous acid, isobutyl pentyl ester | 9.19 | 2.58 |
| 8 | 2-Phenylethyl allyl ether | 10.28 | 0.38 |
| 9 | 12-Hydroxyalliacolide | 12.76 | 8.04 |
| 10 | 7-ethyl-Quinoline | 13.91 | 5.92 |
| ID Compd | Compds | RT (min) | Content μg/g of Dried Extract |
|---|---|---|---|
| 11 | Chlorogenic acid | 13.700 | 395.808 |
| 12 | EGCG | 14.198 | 402.680 |
| 13 | Epicatechin | 14.760 | 576.809 |
| 14 | Epicatechin gallate | 16.230 | 79.513 |
| 15 | Vitexin | 17.785 | 23.197 |
| 16 | Isovitexin | 19.180 | 63.884 |
| 17 | Rutin | 19.603 | 61.581 |
| 18 | Apigetrin | 20.442 | 2481.525 |
| 19 | Myricetin | 20.882 | 221.843 |
| 20 | Quercetin | 22.830 | 487.600 |
| 21 | Apigenin | 24.772 | 184.798 |
| Compounds (Ligands) | Symbols of L-AChE | RMSD (Å) | DS (kcal/mol) |
|---|---|---|---|
| 3-Hydroxydecanoic acid | 1-AChE | 0.82 | −10.1 |
| Propane, 1,1-dipropoxy-(CAS) | 2-AChE | 0.63 | −9.6 |
| (2-(2-butoxyisopropoxy)−2-isopropanol | 3-AChE | 1.72 | −9.8 |
| p-Xylene | 4-AChE | 1.57 | −6.5 |
| Styrene | 5-AChE | 0.93 | −6.3 |
| Oxalic acid, heptyl propyl ester | 6-AChE | 1.11 | −8.8 |
| Sulfurous acid, isobutyl pentyl ester | 7-AChE | 0.81 | −10.3 |
| 2-Phenylethyl allyl ether | 8-AChE | 1.68 | −8.9 |
| 12-Hydroxyalliacolide | 9-AChE | 1.40 | −10.6 |
| 7-ethyl-Quinoline | 10-AChE | 0.82 | −8.6 |
| Chlorogenic acid | 11-AChE | 1.50 | −13.1 |
| EGCG | 12-AChE | 1.24 | −12.5 |
| Epicatechin | 13-AChE | 1.75 | −14.3 |
| Epicatechin gallate | 14-AChE | 1.52 | −13.3 |
| Vitexin | 15-AChE | 1.18 | −12.8 |
| Isovitexin | 16-AChE | 1.09 | −13.8 |
| Rutin | 17-AChE | 1.29 | −14.4 |
| Apigetrin | 18-AChE | 1.03 | −13.9 |
| Myricetin | 19-AChE | 0.77 | −13.1 |
| Quercetin | 20-AChE | 1.51 | −11.0 |
| Apigenin | 21-AChE | 1.26 | −12.3 |
| Berberine chloride | 22-AChE | 1.65 | −12.1 |
| L-AChE Complex | Linkages Number | Amino Acids Interacting with the Ligands [Distance (Å)/E (kcal/mol)/Linkage Type] |
|---|---|---|
| 11-AChE | 5 linkages (2 H-donor, 2 H-acceptor, 1 pi-H) | Tyr70 (2.75/−2.5/H-donor); Glu199 (3.42/−0.6/H-donor); Gly119 (2.78/1.6/H-acceptor); Ser200 (2.68/−1.2/H-acceptor); Asn85 (4.66/−0.6/pi-H) |
| 12-AChE | 4 linkages of H-donor | Ser81 (2.79/−3.7/H-donor); Ser81 (3.18/−2.6/H-donor); Glu199 (3.60/−0.5/H-donor); Glu199 (2.8/−5.0/H-donor) |
| 13-AChE | 4 linkages of H-donor | Ser81 (2.77/−2.8/H-donor); Asn85 (3.10/−1.0/H-donor); Ser200 (2.78/5.6/H-donor); Glu199 (2.99/−2.1/H-donor) |
| 14-AChE | 7 linkages (4 H-donor, 1 H-pi, 2 pi-pi) | His440 (2.59/−1.5/H-donor); Glu199 (2.87/−2.7/H-donor); Tyr70 (2.60/−2.5/H-donor); Asn85 (2.56/−1.9/H-donor); Phe330 (3.91/−1.1/H-pi); Trp84 (3.92/−0.0/pi-pi); Tyr334 (3.74/−0.0/pi-pi) |
| 15-AChE | 4 linkages (2 H-donor, 1 H-acceptor, 1 H-pi) | Asp72 (3.29/−1.0/H-donor); Glu199 (2.84/−4.3/H-donor); Ser200 (2.62/−1.1/H-acceptor); Phe330 (4.32/−1.3/H-pi) |
| 16-AChE | 4 linkages (3 H-donor, 1 H-pi) | Glu199 (3.17/−1.1/H-donor); Glu199 (2.61/−3.5/H-donor); Glu199 (2.89/−4.0/H-donor); Trp84 (3.55/−0.8/H-pi) |
| 17-AChE | 7 linkages (4 H-donor, 2 H-pi, 1 pi-H) | His400 (2.75/−2.2/H-donor); Glu199 (3.00/−2.0/H-donor); Glu199 (2.90/−3.0/H-donor); Trp84 (3.31/−0.9/H-donor); Trp84 (4.02/−0.9/H-pi); Trp84 (4.10/−0.6/H-pi); Asp72 (3.57/−0.8/pi-H) |
| 18-AChE | 3 linkages of H-donor | Glu199 (3.04/−2.7/H-donor); Glu199 (2.92/−3.8/H-donor); Asp72 (2.88/−1.5/H-donor) |
| 19-AChE | 4 linkages of H-donor | Tyr70 (2.98/−2.1/H-donor); Glu199 (2.81/−1.4/H-donor); Glu199 (2.80/−2.1/H-donor); Glu199 (2.84/−1.5/H-donor) |
| 20-AChE | 3 linkages (1 H-donor, 1 H-acceptor, 1 pi-H) | Glu199 (2.75/−5.8/H-donor); His440 (3.02/−1.5/H-acceptor); Gly118 (3.77/−0.7/pi-H) |
| 21-AChE | 3 linkages (1 H-donor, 1 H-acceptor, 1 pi-H) | Glu199 (2.76/−5.8/H-donor); His440 (3.04/−1.1/H-acceptor); Gly118 (3.74/−0.7/pi-H) |
| 22-AChE | 1 H-pi | Tyr121 (4.50/−0.7/H-pi) |
| ID Compd | Mass (Dalton) | Hydrogen Bond Donor | Hydrogen Bond Acceptors | LogP | Molar Refractivity |
|---|---|---|---|---|---|
| 1 | 187.0 | 1 | 3 | 0.848 | 49.01 |
| 2 | 160.0 | 0 | 2 | 22.576 | 46.56 |
| 3 | 190.0 | 1 | 3 | 2.284 | 52.32 |
| 4 | 106.0 | 0 | 0 | 2.303 | 35.92 |
| 5 | 104.0 | 0 | 0 | 2.33 | 36.53 |
| 6 | 230.0 | 0 | 4 | 2.45 | 60.97 |
| 7 | 209.0 | 1 | 3 | 2.787 | 56.26 |
| 8 | 162.0 | 0 | 1 | 2.432 | 51.16 |
| 9 | 187.0 | 1 | 3 | 0.848 | 49.01 |
| 10 | 157.0 | 0 | 1 | 2.797 | 51.210 |
| 11 | 157.0 | 0 | 1 | 2.797 | 51.121 |
| 12 | 353.0 | 5 | 9 | −1.981 | 79.89 |
| 13 | 458.0 | 8 | 11 | 2.233 | 108.92 |
| 14 | 290.0 | 5 | 6 | 1.546 | 72.62 |
| 15 | 442.0 | 7 | 10 | 2.528 | 107.26 |
| 16 | 432.0 | 7 | 10 | −0.066 | 103.53 |
| 17 | 432.0 | 7 | 10 | −0.066 | 103.53 |
| 18 | 610.0 | 10 | 16 | −1.879 | 137.50 |
| 19 | 432.0 | 6 | 10 | −0.107 | 103.54 |
| 20 | 318.0 | 6 | 8 | 1.717 | 75.72 |
| 21 | 302.0 | 5 | 7 | 2.011 | 74.05 |
| 22 | 270.0 | 3 | 5 | 2.420 | 70.81 |
| Lipinski’s rules | ≤500 | ≤5 | ≤10 | ≤5 | 40–130 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, V.B.; Wang, S.-L.; Phan, T.Q.; Doan, M.D.; Phan, T.K.P.; Phan, T.K.T.; Pham, T.H.T.; Nguyen, A.D. Novel Anti-Acetylcholinesterase Effect of Euonymus laxiflorus Champ. Extracts via Experimental and In Silico Studies. Life 2023, 13, 1281. https://doi.org/10.3390/life13061281
Nguyen VB, Wang S-L, Phan TQ, Doan MD, Phan TKP, Phan TKT, Pham THT, Nguyen AD. Novel Anti-Acetylcholinesterase Effect of Euonymus laxiflorus Champ. Extracts via Experimental and In Silico Studies. Life. 2023; 13(6):1281. https://doi.org/10.3390/life13061281
Chicago/Turabian StyleNguyen, Van Bon, San-Lang Wang, Tu Quy Phan, Manh Dung Doan, Thi Kim Phung Phan, Thi Kim Thu Phan, Thi Huyen Thoa Pham, and Anh Dzung Nguyen. 2023. "Novel Anti-Acetylcholinesterase Effect of Euonymus laxiflorus Champ. Extracts via Experimental and In Silico Studies" Life 13, no. 6: 1281. https://doi.org/10.3390/life13061281
APA StyleNguyen, V. B., Wang, S. -L., Phan, T. Q., Doan, M. D., Phan, T. K. P., Phan, T. K. T., Pham, T. H. T., & Nguyen, A. D. (2023). Novel Anti-Acetylcholinesterase Effect of Euonymus laxiflorus Champ. Extracts via Experimental and In Silico Studies. Life, 13(6), 1281. https://doi.org/10.3390/life13061281