Radiomics and Artificial Intelligence for Outcome Prediction in Multiple Myeloma Patients Undergoing Autologous Transplantation: A Feasibility Study with CT Data
Abstract
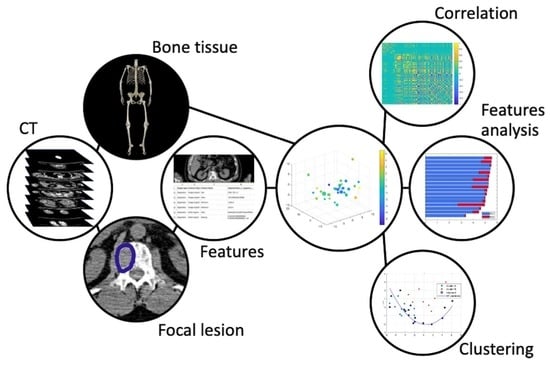
:1. Introduction
2. Materials and Methods
2.1. Study Populations, Inclusion Criteria, and Risk Stratification
2.2. Image Analysis
2.3. Reduction of Redundancy
2.4. Clustering
- Downstream of the PCA process, the two-dimensional feature space given by the two components explaining most of the data variance (namely, PC1 and PC2) was constructed for each data set.
- Given a feature space, the Hough transform of each point in the patient’s set with respect to the family of all parabolas was computed. As this family was characterized by three parameters, i.e., its equation is y_PC2 = ax_PC1^2 + bx_PC1 + c, with a, b, and c being the parameters, and the corresponding parameter space has three dimensions.
- The Hough accumulator was computed by counting the number of times each Hough transform passed through one of the cells of the discretized parameter space.
- The Hough accumulator was filtered by a 5-pixel-side cube centered on the pixel with a maximum grey value. This cube was the smallest one enclosing the cells, with accumulator values higher than 50% of the maximum [32].
3. Results
3.1. Clinical Findings
3.2. AI-Based Analysis
3.3. Feature Ranking
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kasper, D.; Fauci, A.; Hauser, S.; Longo, D.; Jameson, J. Harrison’s Principles of Internal Medicine; McGraw-Hill Education: New York, NY, USA, 2015. [Google Scholar]
- Rajkumar, S.V. Multiple myeloma: Every year a new standard? Hematol Oncol. 2019, 37 (Suppl. 1), 62–65. [Google Scholar] [CrossRef] [Green Version]
- Rajkumar, S.V.; Dimopoulos, M.A.; Palumbo, A.; Blade, J.; Merlini, G.; Mateos, M.V.; Kumar, S.; Hillengass, J.; Kastritis, E.; Richardson, P.; et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014, 15, e538–e548. [Google Scholar] [CrossRef]
- Messiou, C.; Hillengass, J.; Delorme, S.; Lecouvet, F.E.; Moulopoulos, L.; Collins, D.; Blackledge, M.D.; Abildgaard, N.; Østergaard, B.; Schlemmer, H.-P.; et al. Guidelines for acquisition, interpretation, and reporting of whole-body MRI in myeloma: Myeloma response assessment and diagnosis system (MY-RADS). Radiology 2019, 291, 5–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajkumar, S.V. Evolving diagnostic criteria for multiple myeloma. Hematol. Am. Soc. Hematol. Educ. Program. 2015, 2015, 272–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zamagni, E.; Cavo, M.; Fakhri, B.; Vij, R.; Roodman, D. Bones in Multiple Myeloma: Imaging and Therapy. Am. Soc. Clin. Oncol. Educ. Book 2018, 38, 638–646. [Google Scholar] [CrossRef]
- Rasche, L.; Angtuaco, E.J.; Alpe, T.L.; Gershner, G.H.; McDonald, J.E.; Samant, R.S.; Kumar, M.; Van Hemert, R.; Epstein, J.; Deshpande, S.; et al. The presence of large focal lesions is a strong independent prognostic factor in multiple myeloma. Blood 2018, 132, 59–66. [Google Scholar] [CrossRef]
- Siontis, B.; Kumar, S.; Dispenzieri, A.; Drake, M.T.; Lacy, M.Q.; Buadi, F.K.; Dingli, D.; Kapoor, P.; Gonsalves, W.I.; Gertz, M.A.; et al. Positron emission tomography-computed tomography in the diagnostic evaluation of smoldering multiple myeloma: Identification of patients needing therapy. Blood Cancer J. 2015, 5, e364. [Google Scholar] [CrossRef] [Green Version]
- Kosmala, A.; Weng, A.M.; Krauss, B.; Knop, S.; Bley, T.A.; Petritsch, B. Dual-energy CT of the bone marrow in multiple myeloma: Diagnostic accuracy for quantitative differentiation of infiltration patterns. Eur. Radiol. 2018, 28, 5083–5090. [Google Scholar] [CrossRef]
- Pfahler, V.K.A.; D’Anastasi, M.; Dürr, H.; Schinner, R.; Ricke, J.; Baur-Melnyk, A. Tumor Load in Patients with Multiple Myeloma: β₂-Microglobulin Levels Versus Low-Dose Whole-Body CT. Eur. J. Haematol. 2019, 104, 383–389. [Google Scholar] [CrossRef]
- Moulopoulos, L.A.; Koutoulidis, V.; Hillengass, J.; Zamagni, E.; Aquerreta, J.D.; Roche, C.L.; Lentzsch, S.; Moreau, P.; Cavo, M.; Miguel, J.S.; et al. Recommendations for acquisition, interpretation and reporting of whole body low dose CT in patients with multiple myeloma and other plasma cell disorders: A report of the IMWG Bone Working Group. Blood Cancer J. 2018, 8, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filonzi, G.; Mancuso, K.; Zamagni, E.; Nanni, C.; Spinnato, P.; Cavo, M.; Fanti, S.; Salizzoni, E.; Bazzocchi, A. A Comparison of Different Staging Systems for Multiple Myeloma: Can the MRI Pattern Play a Prognostic Role? Am. J. Roentgenol. 2017, 209, 152–158. [Google Scholar] [CrossRef]
- Hillengass, J.; Moulopoulos, L.A.; Delorme, S.; Koutoulidis, V.; Mosebach, J.; Hielscher, T.; Drake, M.; Rajkumar, S.V.; Oestergaard, B.; Abildgaard, N.; et al. Whole-body computed tomography versus conventional skeletal survey in patients with multiple myeloma: A study of the International Myeloma Working Group. Blood Cancer J. 2017, 7, e599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wennmann, M.; Hielscher, T.; Kintzelé, L.; Menze, B.H.; Langs, G.; Merz, M.; Sauer, S.; Kauczor, H.-U.; Schlemmer, H.-P.; Delorme, S.; et al. Spatial Distribution of Focal Lesions in Whole-Body MRI and Influence of MRI Protocol on Staging in Patients with Smoldering Multiple Myeloma According to the New SLiM-CRAB-Criteria. Cancers 2020, 12, 2537. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Paiva, B.; Anderson, K.C.; Durie, B.; Landgren, O.; Moreau, P.; Munshi, N.; Lonial, S.; Bladé, J.; Mateos, M.-V.; et al. International Myeloma Working Group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol. 2016, 17, e328–e346. [Google Scholar] [CrossRef]
- Hillengass, J.; Usmani, S.; Rajkumar, S.V.; Durie, B.G.M.; Mateos, M.-V.; Lonial, S.; Joao, C.; Anderson, K.C.; Garcia-Sanz, R.; Riva, E.; et al. International myeloma working group consensus recommendations on imaging in monoclonal plasma cell disorders. Lancet Oncol. 2019, 20, e302–e312. [Google Scholar] [CrossRef]
- Tagliafico, A.S.; Cea, M.; Rossi, F.; Valdora, F.; Bignotti, B.; Succio, G.; Gualco, S.; Conte, A.; Dominietto, A. Differentiating diffuse from focal pattern on Computed Tomography in multiple myeloma: Added value of a Radiomics approach. Eur. J. Radiol. 2019, 121, 108739. [Google Scholar] [CrossRef]
- Schenone, D.; Lai, R.; Cea, M.; Rossi, F.; Torri, L.; Bignotti, B.; Succio, G.; Gualco, S.; Conte, A.; Dominietto, A.; et al. Radiomics and artificial intelligence analysis of CT data for the identification of prognostic features in multiple myeloma. In Medical Imaging 2020: Computer-Aided Diagnosis; Hahn, H.K., Mazurowski, M.A., Eds.; SPIE: Belingham, WA, USA, 2020; Volume 11314, p. 152. [Google Scholar] [CrossRef] [Green Version]
- Ekert, K.; Hinterleitner, C.; Baumgartner, K.; Fritz, J.; Horger, M. Extended Texture Analysis of Non-Enhanced Whole-Body MRI Image Data for Response Assessment in Multiple Myeloma Patients Undergoing Systemic Therapy. Cancers 2020, 12, 761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morvan, L.; Nanni, C.; Michaud, A.-V.; Jamet, B. Learned Deep Radiomics for Survival Analysis with Attention. In Proceedings of the International Workshop on Predictive Intelligence in Medicine (PRIME 2020), Lima, Peru, 8 October 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 35–45. [Google Scholar]
- Reinert, C.P.; Krieg, E.-M.; Bösmüller, H.; Horger, M. Mid-term response assessment in multiple myeloma using a texture analysis approach on dual energy-CT-derived bone marrow images—A proof of principle study. Eur. J. Radiol. 2020, 131, 109214. [Google Scholar] [CrossRef] [PubMed]
- Jamet, B.; Morvan, L.; Nanni, C.; Michaud, A.-V.; Bailly, C.; Chauvie, S.; Moreau, P.; Touzeau, C.; Zamagni, E.; Bodet-Milin, C.; et al. Random survival forest to predict transplant-eligible newly diagnosed multiple myeloma outcome including FDG-PET radiomics: A combined analysis of two independent prospective European trials. Eur. J. Nucl. Med. Mol. Imaging 2020, 48, 1005–1015. [Google Scholar] [CrossRef]
- Joseph, N.S.; Gentili, S.; Kaufman, J.L.; Lonial, S.; Nooka, A.K. High-risk Multiple Myeloma: Definition and Management. Clin. Lymphoma Myeloma Leuk. 2017, 17S, S80–S87. [Google Scholar] [CrossRef]
- Palumbo, A.; Avet-Loiseau, H.; Oliva, S.; Lokhorst, H.M.; Goldschmidt, H.; Rosinol, L.; Richardson, P.; Caltagirone, S.; Lahuerta, J.J.; Facon, T.; et al. Revised International Staging System for Multiple Myeloma: A Report From International Myeloma Working Group. J. Clin. Oncol. 2015, 33, 2863–2869. [Google Scholar] [CrossRef]
- Fiz, F.; Marini, C.; Piva, R.; Miglino, M.; Massollo, M.; Bongioanni, F.; Morbelli, S.; Bottoni, G.; Campi, C.; Bacigalupo, A.; et al. Adult Advanced Chronic Lymphocytic Leukemia: Computational Analysis of Whole-Body CT Documents a Bone Structure Alteration. Radiology 2014, 271, 805–813. [Google Scholar] [CrossRef] [Green Version]
- Kapur, T.; Pieper, S.; Fedorov, A.; Fillion-Robin, J.-C.; Halle, M.; O’Donnell, L.; Lasso, A.; Ungi, T.; Pinter, C.; Finet, J.; et al. Increasing the impact of medical image computing using community-based open-access hackathons: The NA-MIC and 3D Slicer experience. Med. Image Anal. 2016, 33, 176–180. [Google Scholar] [CrossRef] [Green Version]
- Kikinis, R.; Pieper, S.; Vosburgh, K. 3D Slicer: A Platform for Subject-Specific Image Analysis, Visualization, and Clinical Support. In Intraoperative Imaging and Image-Guided Therapy; Springer: Berlin/Heidelberg, Germany, 2014; Volume 3, pp. 277–289. [Google Scholar] [CrossRef]
- Fedorov, A.; Beichel, R.; Kalpathy-Cramer, J.; Finet, J.; Fillion-Robin, J.-C.; Pujol, S.; Bauer, C.; Jennings, D.; Fennessy, F.; Sonka, M.; et al. 3D Slicer as an Image Computing Platform for the Quantitative Imaging Network. Magn. Reason. Imaging 2012, 30, 1323–1341. [Google Scholar] [CrossRef] [Green Version]
- Jolliffe, I.T. Principal Component Analysis, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2002. [Google Scholar] [CrossRef]
- Bezdek, J.C. Pattern Recognition with Fuzzy Objective Function Algorithms; Kluwer Academic Publishers: Cambridge, MA, USA, 1981. [Google Scholar]
- Beltrametti, M.C.; Massone, A.M.; Piana, M. Hough Transform of Special Classes of Curves. SIAM J. Imaging Sci. 2013, 6, 391–412. [Google Scholar] [CrossRef]
- Massone, A.M.; Perasso, A.; Campi, C.; Beltrametti, M.C. Profile Detection in Medical and Astronomical Images by Means of the Hough Transform of Special Classes of Curves. J. Math. Imaging Vis. 2015, 51, 296–310. [Google Scholar] [CrossRef]
- Roche-Lestienne, C.; Boudry-Labis, E.; Mozziconacci, M.J. Cytogenetics in the management of “chronic myeloid leukemia”: An update by the Groupe francophone de cytogénétique hématologique (GFCH). Ann. Biol. Clin. 2016, 74, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Abe, Y.; Narita, K.; Kitadate, A.; Takeuchi, M.; Matsue, K. Prognostic Significance of Medullary Abnormalities of the Appendicular Skeleton Detected by Low-Dose Whole-Body Multidetector Computed Tomography in Patients with Multiple Myeloma. Blood 2017, 130 (Suppl. 1), 1763. [Google Scholar] [CrossRef]
- Zhovannik, I.; Bussinik, J.; Traverso, A.; Shi, Z.; Kalendralis, P.; Wee, L.; Dekker, A.; Fijiten, R.; Monshouwet, R. Learning from scanners: Bias reduction and feature correction in radiomics. Clin. Transl. Radiat. Oncol. 2019, 16, 33–38. [Google Scholar] [CrossRef] [Green Version]
- Einstein, S.A.; Rong, X.J.; Jensen, C.T.; Liu, X. Quantification and homogenization of image noise between two CT scanner models. J. Appl. Clin. Med. Phys. 2020, 21, 174–178. [Google Scholar] [CrossRef] [Green Version]
- Sollini, M.; Antunovic, L.; Chiti, A.; Kirienko, M. Towards clinical application of image mining: A systematic review on artificial intelligence and radiomics. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 2656–2672. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Characteristic | Number | % |
|---|---|---|
| Patients | 33 | 100 |
| Age (years) Mean | 56 | |
| Age SD 1 | 6.7 | |
| Males | 21 | 66.4 |
| Females | 12 | 34.6 |
| Cytogenetics | ||
| Normal | 22 | 66,7 |
| High risk | 11 | 33,3 |
| Relapsed | 17/33 | 51,5 |
| Days before Relapse (mean) | 1138 | |
| Days of follow-up (mean) | 1317 | |
| International Staging System | ||
| Stage I | 15 | 45.4 |
| Stage II | 9 | 27.3 |
| Stage III | 9 | 27.3 |
| Data Set Name | Vector Dimension | SW Tool | Feature Type | Correlation |
|---|---|---|---|---|
| Data set 1 | 109 | Slicer | focal | no |
| Data set 2 | 8 | Slicer | focal | relapses |
| Data set 3 | 17 | Slicer | focal | global features |
| Method | Data Set | # of Vectors Cluster A | # of Vectors Cluster B | # of Relapses Cluster A | # of Relapses Cluster B |
|---|---|---|---|---|---|
| FCM | 1 | 16 | 17 | 6 | 10 |
| FCM | 2 | 25 | 8 | 8 | 8 |
| FCM | 3 | 23 | 10 | 11 | 5 |
| HTF | 1 | 20 | 13 | 8 | 8 |
| HTF | 2 | 12 | 21 | 7 | 9 |
| HTF | 3 | 25 | 8 | 16 | 0 |
| Method | Data Set | Sensitivity | Specificity | Youden | CSI |
|---|---|---|---|---|---|
| FCM | 1 | 0.46 ± 0.12 | 0.5 ± 0.14 | −0.04 ± 0.13 | 0.3 ± 0.08 |
| FCM | 2 | 0.58 ± 0.35 | 0.55 ± 0.48 | 0.13 ± 0.15 | 0.3 ± 0.08 |
| FCM | 3 | 0.4 ± 0.24 | 0.55 ± 0.22 | −0.06 ± 0.15 | 0.25 ± 0.12 |
| HTF | 1 | 0.38 ± 0.13 | 0.55 ± 0.16 | −0.06 ± 0.15 | 0.25 ± 0.09 |
| HTF | 2 | 0.63 ± 0.19 | 0.33 ± 0.25 | −0.04 ± 0.34 | 0.37 ± 0.16 |
| HTF | 3 | 0.87 ± 0.14 | 0.4 ± 0.13 | 0.27 ± 0.2 | 0.52 ± 0.1 |
| Cytogenetics | 0.45 ± 0.16 | 1.00 ± 0.02 | 0.44 ± 0.16 | 0.44 ± 0.16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schenone, D.; Dominietto, A.; Campi, C.; Frassoni, F.; Cea, M.; Aquino, S.; Angelucci, E.; Rossi, F.; Torri, L.; Bignotti, B.; et al. Radiomics and Artificial Intelligence for Outcome Prediction in Multiple Myeloma Patients Undergoing Autologous Transplantation: A Feasibility Study with CT Data. Diagnostics 2021, 11, 1759. https://doi.org/10.3390/diagnostics11101759
Schenone D, Dominietto A, Campi C, Frassoni F, Cea M, Aquino S, Angelucci E, Rossi F, Torri L, Bignotti B, et al. Radiomics and Artificial Intelligence for Outcome Prediction in Multiple Myeloma Patients Undergoing Autologous Transplantation: A Feasibility Study with CT Data. Diagnostics. 2021; 11(10):1759. https://doi.org/10.3390/diagnostics11101759
Chicago/Turabian StyleSchenone, Daniela, Alida Dominietto, Cristina Campi, Francesco Frassoni, Michele Cea, Sara Aquino, Emanuele Angelucci, Federica Rossi, Lorenzo Torri, Bianca Bignotti, and et al. 2021. "Radiomics and Artificial Intelligence for Outcome Prediction in Multiple Myeloma Patients Undergoing Autologous Transplantation: A Feasibility Study with CT Data" Diagnostics 11, no. 10: 1759. https://doi.org/10.3390/diagnostics11101759
APA StyleSchenone, D., Dominietto, A., Campi, C., Frassoni, F., Cea, M., Aquino, S., Angelucci, E., Rossi, F., Torri, L., Bignotti, B., Tagliafico, A. S., & Piana, M. (2021). Radiomics and Artificial Intelligence for Outcome Prediction in Multiple Myeloma Patients Undergoing Autologous Transplantation: A Feasibility Study with CT Data. Diagnostics, 11(10), 1759. https://doi.org/10.3390/diagnostics11101759