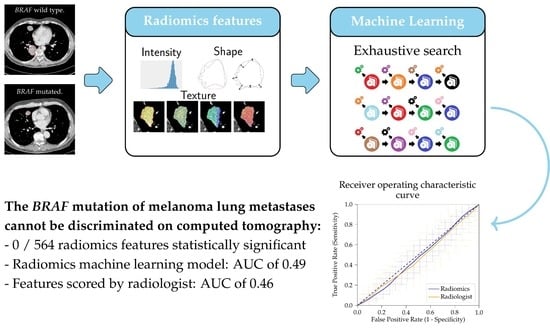
The BRAF P.V600E Mutation Status of Melanoma Lung Metastases Cannot Be Discriminated on Computed Tomography by LIDC Criteria nor Radiomics Using Machine Learning
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Radiomics
2.3. Scoring by Radiologist
2.4. Experimental Setup
2.5. Statistics
3. Results
3.1. Study Population
3.2. Radiomics and LIDC Features and Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- MacKie, R.M.; Hauschild, A.; Eggermont, A.M.M. Epidemiology of invasive cutaneous melanoma. Ann. Oncol. 2009, 20, vi1–vi7. [Google Scholar] [CrossRef] [PubMed]
- Shain, A.H.; Bastian, B.C. From melanocytes to melanomas. Nat. Rev. Cancer 2016, 16, 345–358. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Dyba, T.; Randi, G.; Bettio, M.; Gavin, A.; Visser, O.; Bray, F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur. J. Cancer 2018, 103, 356–387. [Google Scholar] [CrossRef] [PubMed]
- Whiteman, D.C.; Green, A.C.; Olsen, C.M. The Growing Burden of Invasive Melanoma: Projections of Incidence Rates and Numbers of New Cases in Six Susceptible Populations through 2031. J. Investig. Dermatol. 2016, 136, 1161–1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luke, J.J.; Flaherty, K.T.; Ribas, A.; Long, G.V. Targeted agents and immunotherapies: Optimizing outcomes in melanoma. Nat. Rev. Clin. Oncol. 2017, 14, 463–482. [Google Scholar] [CrossRef] [Green Version]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef]
- Curtin, J.A.; Fridlyand, J.; Kageshita, T.; Patel, H.N.; Busam, K.J.; Kutzner, H.; Cho, K.H.; Aiba, S.; Brocker, E.B.; LeBoit, P.E.; et al. Distinct sets of genetic alterations in melanoma. N. Engl. J. Med. 2005, 353, 2135–2147. [Google Scholar] [CrossRef]
- Colombino, M.; Capone, M.; Lissia, A.; Cossu, A.; Rubino, C.; De Giorgi, V.; Massi, D.; Fonsatti, E.; Staibano, S.; Nappi, O.; et al. BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J. Clin. Oncol. 2012, 30, 2522–2529. [Google Scholar] [CrossRef]
- Flaherty, K.T.; Puzanov, I.; Kim, K.B.; Ribas, A.; McArthur, G.A.; Sosman, J.A.; O’Dwyer, P.J.; Lee, R.J.; Grippo, J.F.; Nolop, K.; et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 2010, 363, 809–819. [Google Scholar] [CrossRef] [Green Version]
- Long, G.V.; Stroyakovskiy, D.; Gogas, H.; Levchenko, E.; de Braud, F.; Larkin, J.; Garbe, C.; Jouary, T.; Hauschild, A.; Grob, J.J.; et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N. Engl. J. Med. 2014, 371, 1877–1888. [Google Scholar] [CrossRef] [Green Version]
- Long, G.V.; Flaherty, K.T.; Stroyakovskiy, D.; Gogas, H.; Levchenko, E.; de Braud, F.; Larkin, J.; Garbe, C.; Jouary, T.; Hauschild, A.; et al. Dabrafenib plus trametinib versus dabrafenib monotherapy in patients with metastatic BRAF V600E/K-mutant melanoma: Long-term survival and safety analysis of a phase 3 study. Ann. Oncol. 2017, 28, 1631–1639. [Google Scholar] [CrossRef]
- Larkin, J.; Ascierto, P.A.; Dreno, B.; Atkinson, V.; Liszkay, G.; Maio, M.; Mandala, M.; Demidov, L.; Stroyakovskiy, D.; Thomas, L.; et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N. Engl. J. Med. 2014, 371, 1867–1876. [Google Scholar] [CrossRef] [Green Version]
- Ascierto, P.A.; McArthur, G.A.; Dreno, B.; Atkinson, V.; Liszkay, G.; Di Giacomo, A.M.; Mandala, M.; Demidov, L.; Stroyakovskiy, D.; Thomas, L.; et al. Cobimetinib combined with vemurafenib in advanced BRAF(V600)-mutant melanoma (coBRIM): Updated efficacy results from a randomised, double-blind, phase 3 trial. Lancet Oncol. 2016, 17, 1248–1260. [Google Scholar] [CrossRef]
- Michielin, O.; van Akkooi, A.C.J.; Ascierto, P.A.; Dummer, R.; Keilholz, U.; Committee, E.G. Cutaneous melanoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2019, 30, 1884–1901. [Google Scholar] [CrossRef] [Green Version]
- Cheng, L.; Lopez-Beltran, A.; Massari, F.; MacLennan, G.T.; Montironi, R. Molecular testing for BRAF mutations to inform melanoma treatment decisions: A move toward precision medicine. Mod. Pathol. 2018, 31, 24–38. [Google Scholar] [CrossRef]
- Rios Velazquez, E.; Parmar, C.; Liu, Y.; Coroller, T.P.; Cruz, G.; Stringfield, O.; Ye, Z.; Makrigiorgos, M.; Fennessy, F.; Mak, R.H.; et al. Somatic Mutations Drive Distinct Imaging Phenotypes in Lung Cancer. Cancer Res. 2017, 77, 3922–3930. [Google Scholar] [CrossRef] [Green Version]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Gevaert, O.; Xu, J.; Hoang, C.D.; Leung, A.N.; Xu, Y.; Quon, A.; Rubin, D.L.; Napel, S.; Plevritis, S.K. Non-small cell lung cancer: Identifying prognostic imaging biomarkers by leveraging public gene expression microarray data--methods and preliminary results. Radiology 2012, 264, 387–396. [Google Scholar] [CrossRef]
- Huang, Y.; Liu, Z.; He, L.; Chen, X.; Pan, D.; Ma, Z.; Liang, C.; Tian, J.; Liang, C. Radiomics Signature: A Potential Biomarker for the Prediction of Disease-Free Survival in Early-Stage (I or II) Non-Small Cell Lung Cancer. Radiology 2016, 281, 947–957. [Google Scholar] [CrossRef]
- Nair, V.S.; Gevaert, O.; Davidzon, G.; Napel, S.; Graves, E.E.; Hoang, C.D.; Shrager, J.B.; Quon, A.; Rubin, D.L.; Plevritis, S.K. Prognostic PET 18F-FDG uptake imaging features are associated with major oncogenomic alterations in patients with resected non-small cell lung cancer. Cancer Res. 2012, 72, 3725–3734. [Google Scholar] [CrossRef] [Green Version]
- Parmar, C.; Leijenaar, R.T.; Grossmann, P.; Rios Velazquez, E.; Bussink, J.; Rietveld, D.; Rietbergen, M.M.; Haibe-Kains, B.; Lambin, P.; Aerts, H.J. Radiomic feature clusters and prognostic signatures specific for Lung and Head & Neck cancer. Sci. Rep. 2015, 5, 11044. [Google Scholar]
- Scrivener, M.; de Jong, E.E.C.; van Timmeren, J.E.; Pieters, T.; Ghaye, B.; Geets, X. Radiomics applied to lung cancer: A review. Transl. Cancer Res. 2016, 5, 398–409. [Google Scholar] [CrossRef]
- van Timmeren, J.E.; van Elmpt, W.; Leijenaar, R.T.H.; Reymen, B.; Monshouwer, R.; Bussink, J.; Paelinck, L.; Bogaert, E.; De Wagter, C.; Elhaseen, E.; et al. Longitudinal radiomics of cone-beam CT images from non-small cell lung cancer patients: Evaluation of the added prognostic value for overall survival and locoregional recurrence. Radiother. Oncol. 2019, 136, 78–85. [Google Scholar] [CrossRef]
- Yamamoto, S.; Korn, R.L.; Oklu, R.; Migdal, C.; Gotway, M.B.; Weiss, G.J.; Iafrate, A.J.; Kim, D.W.; Kuo, M.D. ALK molecular phenotype in non-small cell lung cancer: CT radiogenomic characterization. Radiology 2014, 272, 568–576. [Google Scholar] [CrossRef]
- Yang, L.; Dong, D.; Fang, M.; Zhu, Y.; Zang, Y.; Liu, Z.; Zhang, H.; Ying, J.; Zhao, X.; Tian, J. Can CT-based radiomics signature predict KRAS/NRAS/BRAF mutations in colorectal cancer? Eur. Radiol. 2018, 28, 2058–2067. [Google Scholar] [CrossRef]
- Trebeschi, S.; Drago, S.G.; Birkbak, N.J.; Kurilova, I.; Calin, A.M.; Delli Pizzi, A.; Lalezari, F.; Lambregts, D.M.J.; Rohaan, M.W.; Parmar, C.; et al. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Ann. Oncol. 2019, 30, 998–1004. [Google Scholar] [CrossRef] [Green Version]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- Menzer, C.; Menzies, A.M.; Carlino, M.S.; Reijers, I.; Groen, E.J.; Eigentler, T.; de Groot, J.W.B.; van der Veldt, A.A.M.; Johnson, D.B.; Meiss, F.; et al. Targeted Therapy in Advanced Melanoma With Rare BRAF Mutations. J. Clin. Oncol. 2019, 37, 3142–3151. [Google Scholar] [CrossRef] [Green Version]
- Starmans, M.P.; Miclea, R.; van der Voort, S.; Niessen, W.; Thomeer, M.; Klein, S. Classification of Malignant and Benign Liver Tumors Using a Radiomics Approach; SPIE: Bellingham, WA, USA, 2018; Volume 10574. [Google Scholar]
- Starmans, M.P.A.; van der Voort, S.R.; Phil, T.; Klein, S. Workflow for Optimal Radiomics Classification (WORC). Zenodo 2018. Available online: https://github.com/MStarmans91/WORC (accessed on 27 January 2021). [CrossRef]
- Starmans, M.; van der Voort, S.; Vos, M. Fully automatic construction of optimal radiomics workflows. In ECR 2019: Book of Abstracts (B-0908); Insights Imaging; Springer: Vienna, Austria, 2019; Volume 10, p. S379. [Google Scholar] [CrossRef]
- Vos, M.; Starmans, M.P.A.; Timbergen, M.J.M.; van der Voort, S.R.; Padmos, G.A.; Kessels, W.; Niessen, W.J.; van Leenders, G.; Grunhagen, D.J.; Sleijfer, S.; et al. Radiomics approach to distinguish between well differentiated liposarcomas and lipomas on MRI. Br. J. Surg 2019, 106, 1800–1809. [Google Scholar] [CrossRef] [Green Version]
- Martijn, P.A. Starmans. MelaRadiomics. Zenodo 2021. Available online: https://github.com/MStarmans91/MelaRadiomics (accessed on 27 January 2021). [CrossRef]
- Opulencia, P.; Channin, D.S.; Raicu, D.S.; Furst, J.D. Mapping LIDC, RadLex, and lung nodule image features. J. Digit. Imaging 2011, 24, 256–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadeau, C.; Bengio, Y. Inference for the Generalization Error. Mach. Learn. 2003, 52, 239–281. [Google Scholar] [CrossRef] [Green Version]
- Macskassy, S.A.; Provost, F.; Rosset, S. ROC confidence bands: An empirical evaluation. In Proceedings of the 22nd International Conference on Machine Learning, Bonn, Germany, 7–11 August 2005. [Google Scholar]
- Robert, C.; Ribas, A.; Schachter, J.; Arance, A.; Grob, J.J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.M.; Lotem, M.; et al. Pembrolizumab versus ipilimumab in advanced melanoma (KEYNOTE-006): Post-hoc 5-year results from an open-label, multicentre, randomised, controlled, phase 3 study. Lancet Oncol. 2019, 20, 1239–1251. [Google Scholar] [CrossRef]
- Durot, C.; Mule, S.; Soyer, P.; Marchal, A.; Grange, F.; Hoeffel, C. Metastatic melanoma: Pretreatment contrast-enhanced CT texture parameters as predictive biomarkers of survival in patients treated with pembrolizumab. Eur. Radiol. 2019, 29, 3183–3191. [Google Scholar] [CrossRef]
- Sun, R.; Limkin, E.J.; Vakalopoulou, M.; Dercle, L.; Champiat, S.; Han, S.R.; Verlingue, L.; Brandao, D.; Lancia, A.; Ammari, S.; et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: An imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018, 19, 1180–1191. [Google Scholar] [CrossRef]
- Saadani, H.; van der Hiel, B.; Aalbersberg, E.A.; Zavrakidis, I.; Haanen, J.; Hoekstra, O.S.; Boellaard, R.; Stokkel, M.P.M. Metabolic Biomarker-Based BRAFV600 Mutation Association and Prediction in Melanoma. J. Nucl. Med. 2019, 60, 1545–1552. [Google Scholar] [CrossRef]
- Buvat, I.; Orlhac, F. The Dark Side of Radiomics: On the Paramount Importance of Publishing Negative Results. J. Nucl. Med. 2019, 60, 1543–1544. [Google Scholar] [CrossRef]
- Jiangdian, S.; Yanjie, Y. A review of original articles published in the emerging field of radiomics. Eur. J. Radiol. 2020, 127, 108991. [Google Scholar] [CrossRef]
- Timbergen, M.J.M.; Starmans, M.P.A.; Padmos, G.A.; Grünhagen, D.J.; van Leenders, G.J.L.H.; Hanff, D.; Verhoef, C.; Niessen, W.J.; Sleijfer, S.; Klein, S.; et al. Differential diagnosis and mutation stratification of desmoid-type fibromatosis on MRI using radiomics. Eur. J. Radiol. 2020, 131, 109266. [Google Scholar] [CrossRef]
- Starmans, M.P.A.; Timbergen, M.J.; Vos, M.; Renckens, M.; Grünhagen, D.J.; van Leenders, G.J.L.H.; Dwarkasing, R.S.; Willemssen, F.E.J.A.; Niessen, W.J.; Verhoef, C.; et al. Differential diagnosis and molecular stratification of gastrointestinal stromal tumors on CT images using a radiomics approach. arXiv 2020, arXiv:2010.06824. [Google Scholar]
- Starmans, M.P.A.; Buisman, F.E.; Willemssen, F. Prediction of histopathological growth patterns by radiomics and CT-imaging in patients with operable colorectal liver metastases: A proof-of-concept study. In ECR 2020 Book of Abstracts (RPS 1001c-2); Insights Imaging; Springer: Vienna, Austria, 2020; Volume 11, p. 419. [Google Scholar] [CrossRef]
- Castillo, T.J.; Starmans, M.P.; Niessen, W.J.; Schoots, I.; Klein, S.; Veenland, J.F. Classification of Prostate cancer: High grade versus low grade using a radiomics approach. In Proceedings of the IEEE 16th International Symposium on Biomedical Imaging, Venice, Italy, 8–11 April 2019; pp. 1319–1322. [Google Scholar]
- Starmans, M.P.A.; Blazevic, A.; van der Voort, S.R. Prediction of surgery requirement in mesenteric fibrosis on CT using a radiomics approach. In ECR 2019: Book of Abstracts (B-1279); Insights Imaging; Springer: Vienna, Austria, 2019; Volume 10, p. S457. [Google Scholar] [CrossRef]
- Chen, J.; Cheung, H.; Milot, L.; Martel, A.L. AMINN: Autoencoder-based Multiple Instance Neural Network for Outcome Prediction of Multifocal Liver Metastases. arXiv 2020, arXiv:2012.06875. [Google Scholar]
- Manca, A.; Paliogiannis, P.; Colombino, M.; Casula, M.; Lissia, A.; Botti, G.; Caraco, C.; Ascierto, P.A.; Sini, M.C.; Palomba, G.; et al. Mutational concordance between primary and metastatic melanoma: A next-generation sequencing approach. J. Transl. Med. 2019, 17, 289. [Google Scholar] [CrossRef] [Green Version]
- Riveiro-Falkenbach, E.; Villanueva, C.A.; Garrido, M.C.; Ruano, Y.; Garcia-Martin, R.M.; Godoy, E.; Ortiz-Romero, P.L.; Rios-Martin, J.J.; Santos-Briz, A.; Rodriguez-Peralto, J.L. Intra- and Inter-Tumoral Homogeneity of BRAF(V600E) Mutations in Melanoma Tumors. J. Investig. Dermatol. 2015, 135, 3078–3085. [Google Scholar] [CrossRef] [Green Version]
- Valachis, A.; Ullenhag, G.J. Discrepancy in BRAF status among patients with metastatic malignant melanoma: A meta-analysis. Eur. J. Cancer 2017, 81, 106–115. [Google Scholar] [CrossRef]
- Traverso, A.; Wee, L.; Dekker, A.; Gillies, R. Repeatability and Reproducibility of Radiomic Features: A Systematic Review. Int. J. Radiat. Oncol. 2018, 102, 1143–1158. [Google Scholar] [CrossRef] [Green Version]
- Rossi, G.; Barabino, E.; Fedeli, A.; Ficarra, G.; Coco, S.; Russo, A.; Adamo, V.; Buemi, F.; Zullo, L.; Dono, M.; et al. Radiomic Detection of EGFR Mutations in NSCLC. Cancer Res. 2021, 81, 724. [Google Scholar] [CrossRef]



| Patient | BRAF-mt (N = 51) | BRAF-wt (N = 52) | p-Value |
|---|---|---|---|
| Age (years)† | 59 (50–69) | 66 (57–74) | 0.048 |
| Sex | 0.768 | ||
| Male | 25 (49) | 27 (52) | |
| Female | 26 (51) | 25 (48) | |
| Primary tumor localization | 0.027 | ||
| Skin | 49 (96) | 42 (81) | |
| Mucosal | 0 (0) | 6 (11) | |
| Unknown | 2 (4) | 4 (8) | |
| Determination of BRAF-mutation status | 0.851 | ||
| Primary tumor | 9 (18) | 11 (21) | |
| Local recurrence | 1 (2) | 2 (4) | |
| Metastasis | 40 (78) | 39 (75) | |
| Unknown | 1 (2) | 0 (0) | |
| NRAS mutation status $ | Not applicable | ||
| Mutant | - | 22 (42) | |
| Wild type | - | 23 (44) | |
| Unknown | - | 7 (2) | |
| Imaging | |||
| Acquisition protocol | |||
| Slice thickness (mm) †,1 | 1.5 (1.5, 1.5) | 1.5 (1.5, 1.5) | 0.23 |
| Pixel spacing (mm) † | 0.68 (0.64, 0.74) | 0.67 (0.61, 0.73) | 0.16 |
| Tube current (mA) † | 405 (278, 553) | 333 (210, 490) | 0.05 |
| Peak kilovoltage †,1 | 120 (120, 120) | 120 (118, 120) | 0.44 |
| Contrast Agent | 0.84 | ||
| Visipaque 320 | 35 | 37 | |
| Ultravist | 1 | 0 | |
| Omnipaque | 1 | 1 | |
| Optiray | 0 | 1 | |
| Unknown | 14 | 13 | |
| Number of segmented lesions per patient | 0.54 | ||
| One | 20 (39) | 17 (33) | |
| Two | 31 (61) | 35 (67) |
| Model 1 Radiomics All Lesions—WORC | Model 2 Radiomics Largest Lesion | Model 3 Radiomics NRAS Wild Type | Model 4 LIDC All Lesions | Model 5 Radiomics All Lesions—Benchmark | |
|---|---|---|---|---|---|
| AUC | 0.49 [0.38, 0.59] | 0.65 [0.51, 0.79] | 0.49 [0.37, 0.61] | 0.46 [0.38, 0.55] | 0.50 [0.42, 0.58] |
| Accuracy | 0.48 [0.39, 0.57] | 0.61 [0.50, 0.72] | 0.65 [0.58, 0.71] | 0.49 [0.42, 0.56] | 0.50 [0.43, 0.57] |
| Sensitivity | 0.61 [0.44, 0.77] | 0.61 [0.42, 0.80] | 0.94 [0.87, 1.00] | 0.29 [0.11, 0.48] | 0.56 [0.32, 0.80] |
| Specificity | 0.37 [0.22, 0.52] | 0.60 [0.38, 0.82] | 0.08 [0.00, 0.17] | 0.66 [0.46, 0.86] | 0.44 [0.20, 0.69] |
| NPV | 0.53 [0.39, 0.66] | 0.61 [0.46, 0.76] | 0.35 [0.00, 0.75] | 0.52 [0.42, 0.61] | 0.43 [0.21, 0.66] |
| PPV | 0.45 [0.37, 0.53] | 0.63 [0.48, 0.77] | 0.67 [0.62, 0.72] | 0.44 [0.30, 0.58] | 0.47 [0.37, 0.56] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Angus, L.; Starmans, M.P.A.; Rajicic, A.; Odink, A.E.; Jalving, M.; Niessen, W.J.; Visser, J.J.; Sleijfer, S.; Klein, S.; van der Veldt, A.A.M. The BRAF P.V600E Mutation Status of Melanoma Lung Metastases Cannot Be Discriminated on Computed Tomography by LIDC Criteria nor Radiomics Using Machine Learning. J. Pers. Med. 2021, 11, 257. https://doi.org/10.3390/jpm11040257
Angus L, Starmans MPA, Rajicic A, Odink AE, Jalving M, Niessen WJ, Visser JJ, Sleijfer S, Klein S, van der Veldt AAM. The BRAF P.V600E Mutation Status of Melanoma Lung Metastases Cannot Be Discriminated on Computed Tomography by LIDC Criteria nor Radiomics Using Machine Learning. Journal of Personalized Medicine. 2021; 11(4):257. https://doi.org/10.3390/jpm11040257
Chicago/Turabian StyleAngus, Lindsay, Martijn P. A. Starmans, Ana Rajicic, Arlette E. Odink, Mathilde Jalving, Wiro J. Niessen, Jacob J. Visser, Stefan Sleijfer, Stefan Klein, and Astrid A. M. van der Veldt. 2021. "The BRAF P.V600E Mutation Status of Melanoma Lung Metastases Cannot Be Discriminated on Computed Tomography by LIDC Criteria nor Radiomics Using Machine Learning" Journal of Personalized Medicine 11, no. 4: 257. https://doi.org/10.3390/jpm11040257
APA StyleAngus, L., Starmans, M. P. A., Rajicic, A., Odink, A. E., Jalving, M., Niessen, W. J., Visser, J. J., Sleijfer, S., Klein, S., & van der Veldt, A. A. M. (2021). The BRAF P.V600E Mutation Status of Melanoma Lung Metastases Cannot Be Discriminated on Computed Tomography by LIDC Criteria nor Radiomics Using Machine Learning. Journal of Personalized Medicine, 11(4), 257. https://doi.org/10.3390/jpm11040257