First Data on Ornithodoros moubata Aquaporins: Structural, Phylogenetic and Immunogenic Characterisation as Vaccine Targets
Abstract
:1. Introduction
2. Results
2.1. AQP Transcripts Found in O. moubata
2.2. Tissue Expression and Sequence Verification of O. moubata AQPs (OmAQPs)
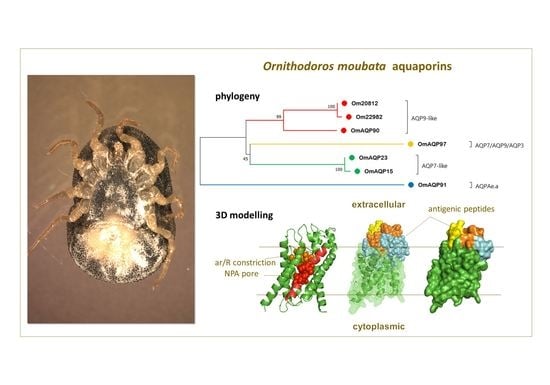
2.3. Phylogenetic Analysis
2.4. OmAQP Structure and 3D Modelling
2.5. Predicted B and T Cell Epitopes and Epitope Exposure; Antigenic Peptide Candidates
2.6. Physicochemical, Allergenic and Toxic Properties of Selected Antigenic Peptides
3. Discussion
4. Materials and Methods
4.1. Selection of Transcripts Containing AQP Coding Sequences
4.2. Ticks and Tick Material
4.3. AQP Sequence Verification and Tissue Expression: PCR Amplification, Cloning and Sequencing
4.4. Prediction and Analysis of the AQP Amino Acid Sequences
4.5. Phylogenetic Analysis
4.6. Protein Structure and Epitope Exposure
4.7. Prediction of B and T Cell Epitopes
4.8. Prediction and Analysis of Antigenic Peptides
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schorderet-Weber, S.; Noack, S.; Selzer, P.M.; Kaminsky, R. Blocking transmission of vector-borne diseases. Int. J. Parasitol. Drugs Drug Resist. 2017, 7, 90–109. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.; Rashid, M.I.; Akbar, H.; Ahmad, L.; Hassan, M.A.; Ashraf, K.; Saeed, K.; Gharbi, M. A systematic review on modelling approaches for economic losses studies caused by parasites and their associated diseases in cattle. Parasitology 2018, 146, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Rochlin, I.; Toledo, A. Emerging tick-borne pathogens of public health importance: A mini-review. J. Med. Microbiol. 2020, 69, 781–791. [Google Scholar] [CrossRef]
- Latif, A.A.; Putterill, J.F.; de Klerk, D.G.; Pienaar, R.; Mans, B.J. Nuttalliella namaqua (Ixodoidea: Nuttalliellidae): First description of the male, immature stages and re-description of the female. PLoS ONE 2012, 7, e41651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Díaz-Martín, V.; Manzano-Román, R.; Oleaga, A.; Pérez-Sánchez, R. New salivary anti-haemostatics containing protective epitopes from Ornithodoros moubata ticks: Assessment of their individual and combined vaccine efficacy. Vet. Parasitol. 2015, 212, 336–349. [Google Scholar] [CrossRef] [Green Version]
- Oleaga, A.; Pérez-Sánchez, R.; Encinas-Grandes, A. Distribution and biology of Ornithodoros erraticus in parts of Spain affected by African swine fever. Vet. Rec. 1990, 126, 32–37. [Google Scholar]
- Mans, B.J.; Neitz, A.W.H. Adaptation of ticks to a blood-feeding environment: Evolution from a functional perspective. Insect Biochem. Mol. Biol. 2004, 34, 1–17. [Google Scholar] [CrossRef]
- Vial, L. Biological and ecological characteristics of soft ticks (Ixodida: Argasidae) and their impact for predicting tick and associated disease distribution. Parasite 2009, 16, 191–202. [Google Scholar] [CrossRef]
- Sonenshine, D.E.; Roe, R.M. Overview: Ticks, People and Animals. In Biology of Ticks; Sonenshine, D.E., Roe, R.M., Eds.; Oxford University Press: Oxford, UK, 2014; Volume 1, pp. 3–16. [Google Scholar]
- Buysse, M.; Duhayon, M.; Cantet, F.; Bonazzi, M.; Duron, O. Vector competence of the African argasid tick Ornithodoros moubata for the Q fever agent Coxiella burnetii. PLoS Negl. Trop. Dis. 2021, 15, e0009008. [Google Scholar] [CrossRef]
- Arias, M.; Jurado, C.; Gallardo, C.; Fernández-Pinero, J.; Sánchez-Vizcaíno, J.M.C. Gaps in African swine fever: Analysis and priorities. Transbound. Emerg. Dis. 2018, 65 (Suppl. S1), 235–247. [Google Scholar] [CrossRef]
- Penrith, M.L.; Bastos, A.D.; Etter, E.M.C.; Beltrán-Alcrudo, D. Epidemiology of African swine fever in Africa today: Sylvatic cycle versus socio-economic imperatives. Transbound. Emerg. Dis. 2019, 66, 672–686. [Google Scholar] [CrossRef] [PubMed]
- Cutler, S.J. Relapsing fever—A forgotten disease revealed. J. Appl. Microbiol. 2010, 108, 1115–1122. [Google Scholar] [CrossRef]
- Talagrand-Reboul, E.; Boyer, P.H.; Bergström, S.; Vial, L.; Boulanger, N. Relapsing fevers: Neglected tick-borne diseases. Front. Cell. Infect. Microbiol. 2018, 8, 98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De la Fuente, J. Controlling ticks and tick-borne diseases looking forward. Ticks Tick Borne Dis. 2018, 9, 1354–1357. [Google Scholar] [CrossRef] [PubMed]
- Ndawula, C., Jr.; Tabor, A.E. Cocktail Anti-Tick Vaccines: The Unforeseen Constraints and Approaches toward Enhanced Efficacies. Vaccines 2020, 8, 457. [Google Scholar] [CrossRef]
- De la Fuente, J.; Kopacek, P.; Lew-Tabor, A.; Maritz-Olivier, C. Strategies for new and improved vaccines against ticks and tick-borne diseases. Parasite Immunol. 2016, 38, 754–769. [Google Scholar] [CrossRef]
- Oleaga, A.; Soriano, B.; Llorens, C.; Pérez-Sánchez, R. Sialotranscriptomics of the argasid tick Ornithodoros moubata along the trophogonic cycle. PLoS Negl. Trop. Dis. 2021, 5, e0009105. [Google Scholar] [CrossRef]
- Pérez-Sánchez, R.; Carnero-Morán, A.; Soriano, B.; Llorens, C.; Oleaga, A. RNA-seq analysis and gene expression dynamics in the salivary glands of the argasid tick Ornithodoros erraticus along the trophogonic cycle. Parasites Vectors 2021, 14, 170. [Google Scholar] [CrossRef]
- Díaz-Martín, V.; Manzano-Román, R.; Valero, L.; Oleaga, A.; Encinas-Grandes, A.; Pérez-Sánchez, R. An insight into the proteome of the saliva of the argasid tick Ornithodoros moubata reveals important differences in saliva protein composition between the sexes. J. Proteom. 2013, 80, 216–235. [Google Scholar] [CrossRef] [Green Version]
- Oleaga, A.; Obolo-Mvoulouga, P.; Manzano-Román, R.; Pérez-Sánchez, R. Functional annotation and analysis of the Ornithodoros moubata midgut genes differentially expressed after blood feeding. Ticks Tick Borne Dis. 2017, 8, 693–708. [Google Scholar] [CrossRef]
- Oleaga, A.; Obolo-Mvoulouga, P.; Manzano-Román, R.; Pérez-Sánchez, R. A proteomic insight into the midgut proteome of Ornithodoros moubata females reveals novel information on blood digestion in argasid ticks. Parasites Vectors 2017, 10, 366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oleaga, A.; Carnero-Morán, A.; Valero, M.L.; Pérez-Sánchez, R. Proteomics informed by transcriptomics for a qualitative and quantitative analysis of the sialoproteome of adult Ornithodoros moubata ticks. Parasites Vectors 2021, 14, 396. [Google Scholar] [CrossRef] [PubMed]
- Obolo-Mvoulouga, P.; Oleaga, A.; Manzano-Román, R.; Pérez-Sánchez, R. Evaluation of the protective efficacy of Ornithodoros moubata midgut membrane antigens selected using omics and in silico prediction algorithms. Ticks Tick Borne Dis. 2018, 9, 1158–1172. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.M.; Ball, A.; Hoppler, S.; Bowman, A.S. Invertebrate aquaporins: A review. J. Comp. Physiol. B 2008, 178, 935–955. [Google Scholar] [CrossRef] [PubMed]
- Gomes, D.; Agasse, A.; Thiébaud, P.; Delrot, S.; Gerós, H.; Chaumont, F. Aquaporins are multifunctional water and solute transporters highly divergent in living organisms. Biochim. Biophys. Acta 2009, 1788, 1213–1228. [Google Scholar] [CrossRef] [Green Version]
- Ni, Z.X.; Cui, J.M.; Zhang, N.Z.; Fu, B.Q. Structural and evolutionary divergence of aquaporins in parasites (Review). Mol. Med. Rep. 2017, 15, 3943–3948. [Google Scholar] [CrossRef] [Green Version]
- Ishibashi, K.; Tanaka, Y.; Morishita, Y. Perspectives on the evolution of aquaporin superfamily. Vitam. Horm. 2020, 112, 1–27. [Google Scholar] [CrossRef]
- Bowman, A.S.; Sauer, J.R. Tick salivary glands: Function, physiology and future. Parasitology 2004, 129, S67–S81. [Google Scholar] [CrossRef]
- Benoit, J.B.; Hansen, I.A.; Szuter, E.M.; Drake, L.L.; Burnett, D.L.; Attardo, G.M. Emerging roles of aquaporins in relation to the physiology of blood-feeding arthropods. J. Comp. Physiol. B 2014, 184, 811–825. [Google Scholar] [CrossRef]
- Guerrero, F.D.; Andreotti, R.; Bendele, K.G.; Cunha, R.C.; Miller, R.J.; Yeater, K.; De León, A.A.P. Rhipicephalus (Boophilus) microplus aquaporin as an effective vaccine antigen to protect against cattle tick infestations. Parasites Vectors 2014, 7, 475. [Google Scholar] [CrossRef] [Green Version]
- Hussein, H.E.; Scoles, G.A.; Ueti, M.W.; Suarez, C.E.; Adham, F.K.; Guerrero, F.D.; Bastos, R.G. Targeted silencing of the Aquaporin 2 gene of Rhipicephalus (Boophilus) microplus reduces tick fitness. Parasites Vectors 2015, 8, 618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ndekezi, C.; Nkamwesiga, J.; Ochwo, S.; Kimuda, M.P.; Mwiine, F.N.; Tweyongyere, R.; Amanyire, W.; Muhanguzi, D. Identification of Ixodid Tick-Specific Aquaporin-1 Potential Anti-tick Vaccine Epitopes: An in-silico Analysis. Front. Bioeng. Biotechnol. 2019, 7, 236. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Sánchez, R.; Manzano-Román, R.; Obolo-Mvoulouga, P.; Oleaga, A. Function-guided selection of midgut antigens from Ornithodoros erraticus ticks and an evaluation of their protective efficacy in rabbits. Vet. Parasitol. 2019, 272, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Scoles, G.A.; Hussein, H.E.; Olds, C.L.; Mason, K.L.; Davis, S.K. Vaccination of cattle with synthetic peptides corresponding to predicted extracellular domains of Rhipicephalus (Boophilus) microplus aquaporin 2 reduced the number of ticks feeding to repletion. Parasites Vectors 2022, 15, 49. [Google Scholar] [CrossRef]
- Campbell, E.M.; Burdin, M.; Hoppler, S.; Bowman, A.S. Role of an aquaporin in the sheep tick Ixodes ricinus: Assessment as a potential control target. Int. J. Parasitol. 2010, 40, 15–23. [Google Scholar] [CrossRef]
- Ball, A.; Campbell, E.M.; Jacob, J.; Hoppler, S.; Bowman, A.S. Identification, functional characterization and expression patterns of a water-specific aquaporin in the brown dog tick, Rhipicephalus sanguineus. Insect. Biochem. Mol. Biol. 2009, 39, 105–112. [Google Scholar] [CrossRef]
- Contreras, M.; de la Fuente, J. Control of infestations by Ixodes ricinus tick larvae in rabbits vaccinated with aquaporin recombinant antigens. Vaccine 2017, 35, 1323–1328. [Google Scholar] [CrossRef]
- Sonenshine, D.E. Excretion and water balance. In Biology of Ticks; Sonenshine, D.E., Roe, R.M., Eds.; Oxford University Press: Oxford, UK, 2014; Volume 1, pp. 206–219. [Google Scholar]
- Nuttall, P.A. Wonders of tick saliva. Ticks Tick-Borne Dis. 2019, 10, 470–481. [Google Scholar] [CrossRef] [Green Version]
- Gaede, K.; Knulle, W. On the mechanism of water vapor sorp- tion from unsaturated atmospheres by ticks. J. Exp. Biol. 1997, 200, 1491–1498. [Google Scholar] [CrossRef]
- Abascal, F.; Irisarri, I.; Zardoya, R. Diversity and evolution of membrane intrinsic proteins. Biochim. Biophys. Acta 2014, 1840, 1468–1481. [Google Scholar] [CrossRef] [Green Version]
- Carbrey, J.M.; Gorelick-Feldman, D.A.; Kozono, D.; Praetorius, J.; Nielsen, S.; Agre, P. Aquaglyceroporin AQP9: Solute permeation and metabolic control of expression in liver. Proc. Natl. Acad. Sci. USA 2013, 100, 2945–2950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caro-Aguilar, I.; Lapp, S.; Pohl, J.; Galinski, M.R.; Moreno, A. Chimeric epitopes delivered by polymeric synthetic linear peptides induce protective immunity to malaria. Microbes Infect. 2005, 7, 1324–1337. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef] [Green Version]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Pierleoni, A.; Martelli, P.L.; Casadio, R. PredGPI: A GPI-anchor predictor. BMC Bioinform. 2008, 9, 392. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.; Brunak, S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput. 2002, 7, 310–322. [Google Scholar]
- Steentoft, C.; Vakhrushev, S.Y.; Joshi, H.J.; Kong, Y.; Vester-Christensen, M.B.; Schjoldager, K.T.; Lavrsen, K.; Dabelsteen, S.; Pedersen, N.B.; Marcos-Silva, L.; et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013, 32, 1478–1488. [Google Scholar] [CrossRef] [Green Version]
- Doytchinova, I.A.; Flower, D.R. Bioinformatic Approach for Identifying Parasite and Fungal Candidate Subunit Vaccines. Open Vaccine J. 2008, 1, 22–26. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11.0. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Möller, S.; Croning, M.D.; Apweiler, R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 2001, 17, 646–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emini, E.A.; Hughes, J.V.; Perlow, D.S.; Boger, J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J. Virol. 1985, 55, 836–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef] [Green Version]
- Chou, P.Y.; Fasman, G.D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv. Enzymol. Relat. Areas Mol. Biol. 1978, 47, 45–148. [Google Scholar] [CrossRef]
- Karplus, P.A.; Schulz, G.E. Prediction of chain flexibility in proteins. Naturwissenschaften 1985, 72, 212–213. [Google Scholar] [CrossRef]
- Parker, J.M.; Guo, D.; Hodges, R.S. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: Correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry 1986, 25, 5425–5432. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Wiederstein, M.; Sippl, J.S. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [Green Version]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.; Thornton, J.M.S. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Schrödinger, L.; DeLano, W. PyMOL. 2020. Available online: http://www.pymol.org/pymol (accessed on 25 October 2021).
- Kolaskar, A.; Tongaonkar, P. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990, 276, 172–174. [Google Scholar] [CrossRef] [Green Version]
- Larsen, J.E.P.; Lund, O.; Nielsen, M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006, 2, 2. [Google Scholar] [CrossRef] [Green Version]
- Saha, S.; Raghava, G.P.S. Prediction of Continuous B-cell Epitopes in an Antigen Using Recurrent Neural Network. Proteins 2006, 65, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Raghava, G.P.S. BcePred:Prediction of Continuous B-Cell Epitopes in Antigenic Sequences Using Physico-chemical Properties. In Proceedings of the International Conference on Artificial Immune Systems, Taormina, Italy, 28–31 August 2012; Nicosia, G., Cutello, V., Bentley, P.J., Timis, J., Eds.; ICARIS, LNCS; Springer: Cham, Switzerland, 2004; Volume 3239, pp. 197–204. [Google Scholar]
- Ponomarenko, J.V.; Bui, H.; Li, W.; Fusseder, N.; Bourne, P.E.; Sette, A.; Peters, B. ElliPro: A new structure-based tool for the prediction of antibody epitopes. BMC Bioinform. 2008, 9, 514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, M.; Lundegaard, C.; Lund, O.; Keçsmir, C. The role of the proteasome in generating cytotoxic T-cell epitopes: Insights obtained from improved predictions of proteasomal cleavage. Immunogenetics 2005, 57, 33–41. [Google Scholar] [CrossRef]
- Hebditch, M.; Carballo-Amador, M.A.; Charonis, S.; Curtis, R.; Warwicker, J. Protein-Sol: A web tool for predicting protein solubility from sequence. Bioinformatics 2017, 33, 3098–3100. [Google Scholar] [CrossRef] [Green Version]
- Kurgan, L.; Razib, A.A.; Aghakhani, S.; Dick, S.; Mizianty, M.; Jahandideh, S. CRYSTALP2: Sequence-based protein crystallization propensity prediction. BMC Struct. Biol. 2009, 9, 50. [Google Scholar] [CrossRef] [Green Version]
- Dimitrov, I.; Naneva, L.; Doytchinova, I.; Bangov, I. AllergenFP: Allergenicity prediction by descriptor fingerprints. Bioinformatics 2014, 30, 846–851. [Google Scholar] [CrossRef]
- Dimitrov, I.; Bangov, I.; Flower, D.R.; Doytchinova, I. AllerTOP v.2—A server for in silico prediction of allergens. J. Mol. Model. 2014, 20, 2278. [Google Scholar] [CrossRef]
- Maurer-Stroh, S.; Krutz, N.; Kern, P.; Gunalan, V.; Nguyen, M.; Limviphuvadh, V.; Eisenhaber, F.; Gerberick, G. AllerCatPro—Prediction of protein allergenicity potential from the protein sequence. Bioinformatics 2019, 35, 3020–3027. [Google Scholar] [CrossRef] [Green Version]
- Chaudhary, K.; Kumar, R.; Singh, S.; Tuknait, A.; Gautam, A.; Mathur, D.; Anand, P.; Varshney, G.C.; Raghava, G.P.S. A web server and mobile app for computing hemolytic potency of peptides. Sci. Rep. 2016, 6, 22843. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Raghava, G.P.S. In Silico Approach for Predicting Toxicity of Peptides and Proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ettayapuram Ramaprasad, A.S.; Singh, S.; Gajendra, P.S.R.; Venkatesan, S. AntiAngioPred: A Server for Prediction of Anti-Angiogenic Peptides. PLoS ONE 2015, 10, e0136990. [Google Scholar] [CrossRef]




| List | Transcript Dataset | Transcript Code | Protein Accession (GenBank/Uniprot) | Name | ORF Length (bp) | Match_Sequence | Species | Protein Name |
|---|---|---|---|---|---|---|---|---|
| 1 | M | ci|000144090 | - | OmAQP90 | 867 | XP_029845132.1 | I. scapularis | aquaporin-9 isoform X1 |
| 2 | SG | OM_20812 | MBZ3958194 | Om20812 | 843 | |||
| 3 | SG | OM_22982 | MBZ3960076 | Om22982 | 882 | |||
| 4 | M, SG | ci|000124891, OM_7339 | A0A1Z5L1C7 | OmAQP91 | 816 | XP_029833586.1 | I. scapularis | aquaporin AQPAe.a-like |
| 5 | M | ci|000113997 | A0A1Z5L6U6 | OmAQP97 | 879 | CAX48963.1 | R. sanguineus | aquaglyceroporin |
| 6 | M, SG | ci|000114723, OM_21119 | A0A1Z5L547 | OmAQP23 | 849 | CAR66115.1 | R. sanguineus | water-specific aquaporin |
| 7 | M | ci|000148315 | A0A1Z5KVQ3 | OmAQP15 | 915 |
| List | Name | Tissue Expression | GenBank/Uniprot Code | Protein Length (aa) | MW (kDa) | pI | Signal P | GPI Anchor | N-glycosilation Sites | O-glycosilation Sites | VaxiJen Score |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | OmAQP90 | SG, M, CG | - | 288 | 31.2 | 6.4 | no | no | no | 128T, 132S | 0.5979 |
| 2 | Om20812 | SG, M | MBZ3958194 | 280 | 30.1 | 5.4 | no | no | no | 126T, 128T | 0.6173 |
| 3 | Om22982 | SG, M | MBZ3960076 | 293 | 31.4 | 5.1 | no | no | no | 126T, 128T | 0.6597 |
| 4 | OmAQP91 | SG, M | A0A1Z5L1C7 | 271 | 28.4 | 6.0 | no | no | no | no | 0.3729 |
| 5 | OmAQP97 | SG, M, CG | A0A1Z5L6U6 | 292 | 32.6 | 7.2 | no | no | no | no | 0.5386 |
| 6 | OmAQP23 | SG, M, CG | A0A1Z5L547 | 282 | 30.3 | 7.7 | no | no | no | no | 0.5234 |
| 7 | OmAQP15 | SG, M | A0A1Z5KVQ3 | 304 | 32.7 | 7.6 | no | no | no | no | 0.5007 |
| List | Extracellular Domain | Clade | AQPs in the Clade | Peptide Name | Peptide Sequence | Peptide Length | Peptide Position | Peptide Type |
|---|---|---|---|---|---|---|---|---|
| 1 | Loop A | AQP9-like | OmAQP90 Om20812 Om22982 | OmAQP90_A Om20812_A Om22982_A | AGRQEENGH AGRQEHNAG AGRQEHNAG | 9 9 9 | 36–44 34–42 34–42 | B B B |
| 2 | ||||||||
| 3 | ||||||||
| 4 | AQP7-like | OmAQP23 OmAQP15 | OmAQP23_A OmAQP15_A | KFDRAGNIGYA KFDRAGNIGYA | 11 11 | 38–48 38–48 | B, T B, T | |
| 5 | ||||||||
| 6 | AQP7/AQP9/AQP3 | OmAQP97 | OmAQP97_A | HYIFSGQKD | 9 | 42–50 | B | |
| 7 | AQPAe.a | OmAQP91 | OmAQP91_A | CGTCTNWGRGGEPSIA | 16 | 42–56 | B, T | |
| 8 | Loop C | AQP9-like | OmAQP90 Om20812 Om22982 | OmAQP90_C Om20812_C Om22982_C | RDAINMFDGGVRSVVGPTGTASIFSTYPREG RDAIDAVDSGVRSVLGPTGTAPIFATYPREG RDAIDAVDSGVRSVLGPTGTAPIFATYPREG | 31 31 31 | 111–141 109–139 109–139 | B, T B, T B, T |
| 9 | ||||||||
| 10 | ||||||||
| 11 | AQP7-like | OmAQP23 OmAQP15 | OmAQP23_C OmAQP15_C | KGAFDNYDGGFRATTGVNGTADVFASYPRDF KGAFDNYDGGFRATTGVNGTADVFASYPRDF | 31 31 | 115–145 115–145 | B, T B, T | |
| 12 | ||||||||
| 13 | AQP7/AQP9/AQP3 | OmAQP97 | OmAQP97_C | NYIDALDHYDGGERQIFGDRGTGILLTTFPNEH | 33 | 117–149 | B, T | |
| 14 | AQPAe.a | OmAQP91 | OmAQP91_C | AVTPEERQGLLGGTALSEGVTPFQG | 25 | 120–144 | B, T | |
| 15 | Loop E | AQP9-like | OmAQP90 Om20812 Om22982 | OmAQP90_E Om20812_E Om22982_E | SYNCMAALNPARDIGPRVFTAVAGWGSEVFSFRNYQ SYNCMAPLNPARDLGPRVFTAIAGWGMEVFSVRDY SYNCMAPLNPARDLGPRVFTAIAGWGMEVFSVRDY | 36 35 35 | 194–229 193–227 193–227 | B, T B, T B, T |
| 16 | ||||||||
| 17 | ||||||||
| 18 | AQP7-like | OmAQP23 OmAQP15 | OmAQP23_E OmAQP15_E | PLNPARDLGPRIFTAMAGWGTEVFSFRDYN PLNPARDLGPRIFTAMAGWGTEVFSFRDYN | 30 30 | 204–233 204–233 | B, T B, T | |
| 19 | ||||||||
| 20 | AQP7/AQP9/AQP3 | OmAQP97 | OmAQP97_E | NPARDFPPRVLASIVGYGPEVFTYRH | 26 | 210–235 | B, T | |
| 21 | AQPAe.a | OmAQP91 | OmAQP91_E | ASMNTARTFGPAVISGAFDDH | 21 | 195–215 | B, T |
| List | Transcript Code | Primer Name | Primer Sequence (5′-3′) | Product Size (bp) | Tm |
|---|---|---|---|---|---|
| 1 | ci|000144090 | OmAQP90F OmAQP90R | ATGAAGGTGTACATTCGGAGTC CTAGATGTTGGTTGTCTCTTTGG | 867 | 57 °C |
| 2 | OM_20812 | Om20812_22982F Om20812R | ATGAAGATACAGAGCACATTCGTC TCACAGCCAAGTTGGGCCTA | 843 | 60 °C |
| 3 | OM_22982 | Om20812_22982F Om22982R | ATGAAGATACAGAGCACATTCGTC CTATACACAACTATCGCAGCTGAAT | 882 | 60 °C |
| 4 | ci|000124891 (OM_7339) | OmAQP91F OmAQP91R | ATGGGCCGTGTTCGCCAATT TCAGATGGCCGTGGTGCG | 816 | 62.8 °C |
| 5 | ci|000113997 | OmAQP97F OmAQP97R | ATGGCAAACCCACCGTTCC TTAGACACCGGTCTTTTCTGTCC | 879 | 61 °C |
| 6 | ci|000114723 (OM_21129) | OmAQP23_15F OmAQP23R | ATGATTCTGGATAAAGTGAAGATTAAG TCATTCGAGGGAATACCCAC | 849 | 59 °C |
| 7 | ci|000148315 | OmAQP23_15F OmAQP15R | ATGATTCTGGATAAAGTGAAGATTAAG CTAGACCTTCGACTGTTTTTCGT | 915 | 59 °C |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Sánchez, R.; Cano-Argüelles, A.L.; González-Sánchez, M.; Oleaga, A. First Data on Ornithodoros moubata Aquaporins: Structural, Phylogenetic and Immunogenic Characterisation as Vaccine Targets. Pathogens 2022, 11, 694. https://doi.org/10.3390/pathogens11060694
Pérez-Sánchez R, Cano-Argüelles AL, González-Sánchez M, Oleaga A. First Data on Ornithodoros moubata Aquaporins: Structural, Phylogenetic and Immunogenic Characterisation as Vaccine Targets. Pathogens. 2022; 11(6):694. https://doi.org/10.3390/pathogens11060694
Chicago/Turabian StylePérez-Sánchez, Ricardo, Ana Laura Cano-Argüelles, María González-Sánchez, and Ana Oleaga. 2022. "First Data on Ornithodoros moubata Aquaporins: Structural, Phylogenetic and Immunogenic Characterisation as Vaccine Targets" Pathogens 11, no. 6: 694. https://doi.org/10.3390/pathogens11060694
APA StylePérez-Sánchez, R., Cano-Argüelles, A. L., González-Sánchez, M., & Oleaga, A. (2022). First Data on Ornithodoros moubata Aquaporins: Structural, Phylogenetic and Immunogenic Characterisation as Vaccine Targets. Pathogens, 11(6), 694. https://doi.org/10.3390/pathogens11060694