Isolation of Naegleria spp. from a Brazilian Water Source
Abstract
:1. Introduction
2. Results
2.1. Limnologic Characterization
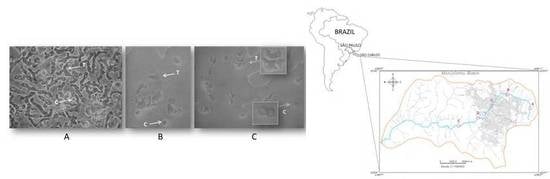
2.2. Light Microscopy
2.3. PCR Amplification and Sequence Analysis
2.4. Phylogeny
3. Discussion
3.1. Eutrophication along the Monjolinho River
3.2. Presence of Naegleria Thermophilic Species in Brazil
3.3. Naegleria spp. Diversity in Brazil
4. Materials and Methods
4.1. Description of the Geographical Area and Sample Collection
4.2. Limnology and Sampling Process
4.3. Amoeba Isolation and Culturing
4.4. Morphological Characterization
4.5. DNA Extraction, Amplification and Sequencing
4.6. Phylogenetic Inferences
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kilvington, S.; Beeching, J. Identification and epidemiological typing of Naegleria fowleri with DNA probes. Appl. Environ. Microbiol. 1995, 61, 2071–2078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latifi, A.R.; Niyyati, M.; Lorenzo-Morales, J.; Haghighi, A.; Tabaei, S.J.S.; Lasjerdi, Z.; Azargashb, E. Occurrence of Naegleria species in therapeutic geothermal water sources, Northern Iran. Acta Parasitol. 2017, 62, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Trabelsi, H.; Dendana, F.; Sellami, A.; Sellami, H.; Cheikhrouhou, F.; Neji, S.; Makni, F.; Ayadi, A. Pathogenic free-living amoebae: Epidemiology and clinical review. Pathol. Boil. 2012, 60, 399–405. [Google Scholar] [CrossRef] [PubMed]
- De Jonckheere, J.F. What do we know by now about the genus Naegleria? Exp. Parasitol. 2014, 145. [Google Scholar] [CrossRef]
- Opperdoes, F.R.; De Jonckheere, J.F.; Tielens, A.G. Naegleria gruberi metabolism. Int. J. Parasitol. 2011, 41, 915–924. [Google Scholar] [CrossRef]
- Fritz-Laylin, L.K.; Prochnik, S.E.; Ginger, M.L.; Dacks, J.B.; Carpenter, M.L.; Field, M.C.; Kuo, A.; Paredez, A.; Chapman, J.; Pham, J.; et al. The Genome of Naegleria gruberi Illuminates Early Eukaryotic Versatility. Cell 2010, 140, 631–642. [Google Scholar] [CrossRef] [Green Version]
- Cogo, P.E.; Scaglia, M.; Gatti, S.; Rossetti, F.; Alaggio, R.; Laverda, A.M.; Zhou, L.; Xiao, L.; Visvesvara, G.S. Fatal Naegleria fowleri Meningoencephalitis, Italy. Emerg. Infect. Dis. 2004, 10, 1835–1837. [Google Scholar] [CrossRef]
- Majid, M.A.A.; Mahboob, T.; Mong, B.G.J.; Jaturas, N.; Richard, R.L.; Tian-Chye, T.; Phimphila, A.; Mahaphonh, P.; Aye, K.N.; Aung, W.L.; et al. Pathogenic waterborne free-living amoebae: An update from selected Southeast Asian countries. PLoS ONE 2017, 12, 1–17. [Google Scholar]
- Tung, M.-C.; Hsu, B.-M.; Tao, C.-W.; Lin, W.-C.; Tsai, H.-F.; Ji, D.-D.; Shen, S.-M.; Chen, J.-S.; Shih, F.-C.; Huang, Y.-L. Identification and significance of Naegleria fowleri isolated from the hot spring which related to the first primary amebic meningoencephalitis (PAM) patient in Taiwan. Int. J. Parasitol. 2013, 43, 691–696. [Google Scholar] [CrossRef]
- Sukthana, Y.; Lekkla, A.; Sutthikornchai, C.; Wanapongse, P.; Vejjajiva, A.; Bovornkitti, S. Spa, springs and safety. Southeast Asian J. Trop. Med. Public Heal. 2005, 36, 10–16. [Google Scholar]
- De Jonckheere, J.F. The impact of man on the occurrence of the pathogenic free-living amoeboflagellateNaegleria fowleri. Futur. Microbiol. 2012, 7, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Tiewcharoen, S.; Phurttikul, W.; Rabablert, J.; Auewarakul, P.; Roytrakul, S.; Chetanachan, P.; Atithep, T.; Junnu, V. Effect of synthetic antimicrobial peptides on Naegleria fowleri trophozoites. Southeast Asian J. Trop. Med. Public Heal. 2014, 45, 537–546. [Google Scholar]
- Schuster, F. Opportunistic amoebae: challenges in prophylaxis and treatment. Drug Resist. Updat. 2004, 7, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Adl, S.M.; Simpson, A.G.B.; Farmer, M.A.; Andersen, R.A.; Anderson, O.R.; Barta, J.R.; Bowser, S.S.; Brugerolle, G.; Fensome, R.A.; Fredericq, S.; et al. The New Higher Level Classification of Eukaryotes with Emphasis on the Taxonomy of Protists. J. Eukaryot. Microbiol. 2005, 52, 399–451. [Google Scholar] [CrossRef] [PubMed]
- Mann, D.G.; Hoppenrath, M.; Cárdenas, P.; Lukeš, J.; Powell, M.J.; Shimano, S.; Bass, D.; Shadwick, L.; del Campo, J.; James, T.Y.; et al. Revisions to the Classification, Nomenclature, and Diversity of Eukaryotes. J. Eukaryot. Microbiol. 2019, 66, 4–119. [Google Scholar]
- De Jonckheere, J.F. A century of research on the amoeboflagellate genus Naegleria. Acta Protozool. 2002, 41, 309–342. [Google Scholar]
- Delafont, V.; Rodier, M.-H.; Maisonneuve, E.; Cateau, E. Vermamoeba vermiformis: a Free-Living Amoeba of Interest. Microb. Ecol. 2018, 76, 991–1001. [Google Scholar] [CrossRef]
- Biswal, D.S.R.S.D.A.K.A. Role of Acanthamoeba in Granulomatous Encephalitis: A Review. J. Infect. Dis. Immune Ther. 2017, 1, 1–12. [Google Scholar]
- Visvesvara, G.S. Amebic meningoencephalitides and keratitis: challenges in diagnosis and treatment. Curr. Opin. Infect. Dis. 2010, 23, 590–594. [Google Scholar] [CrossRef]
- Qvarnstrom, Y.; Silva, A.J.; Schuster, F.L.; Gelman, B.B.; Visvesvara, G.S. Molecular Confirmation of Sappinia pedata as a Causative Agent of Amoebic Encephalitis. J. Infect Dis. 2018, 199, 1139–1142. [Google Scholar] [CrossRef] [Green Version]
- Khan, N.A.; Ong, T.Y.Y.; Siddiqui, R. Targeting Brain-Eating Amoebae Infections. ACS Chem. Neurosci. 2017, 8, 687–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Filippo, M.M.; Berrilli, F.; Di Cave, D.; Novelletto, A. Novel data from Italian Vermamoeba vermiformis isolates from multiple sources add to genetic diversity within the genus. Parasitol. Res. 2019, 118, 1751–1759. [Google Scholar] [CrossRef] [PubMed]
- Ong, T.Y.Y.; Khan, N.A.; Siddiqui, R. Brain-Eating Amoebae: Predilection Sites in the Brain and Disease Outcome. J. Clin. Microbiol. 2017, 55, 1989–1997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niyyati, M.; Dodangeh, S.; Lorenzo-Morales, J. A Review of the Current Research Trends in the Application of Medicinal Plants as a Source for Novel Therapeutic Agents Against Acanthamoeba Infections. Heal. Serv. Iran. J. Pharm. Res. 2016, 15, 893–900. [Google Scholar]
- De Jonckheere, J.F. Origin and evolution of the worldwide distributed pathogenic amoeboflagellate Naegleria fowleri. Infect. Genet. Evol. 2011, 11, 1520–1528. [Google Scholar] [CrossRef] [PubMed]
- Debnath, A.; Tunac, J.B.; Galindo-Gómez, S.; Silva-Olivares, A.; Shibayama, M.; McKerrow, J.H. Corifungin, a New Drug Lead against Naegleria, Identified from a High-Throughput Screen. Antimicrob. Agents Chemother. 2012, 56, 5450–5457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoder, J.S.; Eddy, B.A.; Visvesvara, G.S.; Capewell, L.; Beach, M.J. The epidemiology of primary amoebic meningoencephalitis in the USA, 1962-2008. Epidemiol. Infect. 2010, 138, 968–975. [Google Scholar] [CrossRef] [Green Version]
- Visvesvara, G.S. Free-Living Amebae as Opportunistic Agents of Human Disease. J. Neuroparasitology 2010, 1, 1–13. [Google Scholar] [CrossRef]
- Bellini, N.K.; Santos, T.M.; Da Silva, M.T.A.; Thiemann, O.H. The therapeutic strategies against Naegleria fowleri. Exp. Parasitol. 2018, 187, 1–11. [Google Scholar] [CrossRef]
- Salazar, H.; Moura, H.; Fernandes, O.; Peralta, J. Isolation of Naegleria fowleri from a lake in the city of Rio de Janeiro, Brazil. Trans. R. Soc. Trop. Med. Hyg. 1986, 80, 348–349. [Google Scholar] [CrossRef]
- Aparecida, M.; Aristeu, J. Isolation of potencially pathogenic free-living amoebas in hospital dust. Rev Saude Publ. 2003, 37, 242–246. [Google Scholar]
- Carlesso, A.M.; Simonetti, A.B.; Artuso, G.L.; Rott, M.B. [Isolation and identification of potentially pathogenic free-living amoebae in samples from environments in a public hospital in the city of Porto Alegre, Rio Grande do Sul]. Rev. da Soc. Bras. de Med. Trop. 2007, 40, 316–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maciver, S.K.; Piñero, J.E.; Lorenzo-Morales, J. Is Naegleria fowleri an Emerging Parasite? Trends Parasitol. 2020, 36, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, L.H.; Rocha, S.; Pinto, R.M.F.; Caseiro, M.M.; Costa, S.O.P. Prevalence of potentially pathogenic free-living amoebae from Acanthamoeba and Naegleria genera in non-hospital, public, internal environments from the city of Santos, Brazil. Braz. J. Infect. Dis. 2009, 13, 395–397. [Google Scholar] [CrossRef] [Green Version]
- Pimentel, L.A.; Dantas, A.F.M.; Uzal, F.; Riet-Correa, F. Meningoencephalitis caused by Naegleria fowleri in cattle of northeast Brazil. Res. Veter- Sci. 2012, 93, 811–812. [Google Scholar] [CrossRef]
- Henker, L.C.; Da Cruz, R.A.S.; Da Silva, F.S.; Driemeier, D.; Sonne, L.; Uzal, F.A.; Pavarini, S.P. Meningoencephalitis due to Naegleria fowleri in cattle in southern Brazil. Revista Brasileira de Parasitologia Veterinária 2019, 28, 514–517. [Google Scholar] [CrossRef] [Green Version]
- Edagawa, A.; Kimura, A.; Kawabuchi-Kurata, T.; Kusuhara, Y.; Karanis, P. Isolation and genotyping of potentially pathogenic Acanthamoeba and Naegleria species from tap-water sources in Osaka, Japan. Parasitol. Res. 2009, 105, 1109–1117. [Google Scholar] [CrossRef]
- El-Badry, A.A.; Aufy, S.M.; El-Wakil, E.S.; Rizk, E.M.; Mahmoud, S.S.; Taha, N.Y. First identification of Naegleria species and Vahlkampfia ciguana in Nile water, Cairo, Egypt: Seasonal morphology and phylogenetic analysis. J. Microbiol. Immunol. Infect. 2018. [Google Scholar] [CrossRef]
- Ithoi, I.; Ahmad, A.F.; Nissapatorn, V.; Lau, Y.L.; Mahmud, R.; Mak, J.W. Detection of Naegleria Species in Environmental Samples from Peninsular Malaysia. PLOS ONE 2011, 6, e24327. [Google Scholar] [CrossRef] [Green Version]
- Pélandakis, M.; Pernin, P. Use of Multiplex PCR and PCR Restriction Enzyme Analysis for Detection and Exploration of the Variability in the Free-Living Amoeba Naegleria in the Environment. Appl. Environ. Microbiol. 2002, 68, 2061–2065. [Google Scholar] [CrossRef] [Green Version]
- Pelandakis, M.; Serre, S.; Pernin, P. Analysis of the 5.8S rRNA gene and the internal transcribed spacers in Naegleria spp. and in N. fowleri. J. Eukaryot. Microbiol. 2000, 47, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Junk, W.J.; Piedade, M.T.F.; Lourival, R.; Wittmann, F.; Kandus, P.; Lacerda, L.D.; Bozelli, R.L.; Esteves, F.A.; Nunes da Cunha, C.; Maltchik, L.; et al. Brazilian wetlands: Their definition, delineation, and classification for research, sustainable management, and protection. Aquat. Conserv. Mar. Freshw. Ecosyst. 2014, 24, 5–22. [Google Scholar] [CrossRef]
- Barrilli, G.H.C.; Rocha, O.; Negreiros, N.F.; Verani, J.R. Influence of environmental quality of the tributaries of the Monjolinho River on the relative condition factor (Kn) of the local ichthyofauna. Biota Neotropica 2015, 15, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Campagna, A.F.; Fracácio, R.; Rodrigues, B.K.; Eler, M.N.; Verani, N.F.; Espíndola, E.L.G. Analyses of the sediment toxicity of Monjolinho River, São Carlos, São Paulo State, Brazil, using survey, growth and gill morphology of two fish species (Danio rerio and Poecilia reticulata). Braz. Arch. Boil. Technol. 2008, 51, 193–201. [Google Scholar] [CrossRef]
- Buhse, H.E.; Page, F.C. A New Key to Freshwater and Soil Gymnamoeba. Trans. Am. Microsc. Soc. 1988, 107, 379. [Google Scholar] [CrossRef]
- Yang, X.-E.; Wu, X.; Hao, H.-L.; He, Z.-L. Mechanisms and assessment of water eutrophication. J. Zhejiang Univ. Sci. B 2008, 9, 197–209. [Google Scholar] [CrossRef]
- Baird, C.; Cann, M. Environmental Chemistry, 5th ed.; W. H. Freeman, Macmillan: New York, NY, USA, 2012. [Google Scholar]
- Brasil. Conselho Nacional do Meio Aambiente Resolução CONAMA n 357; Brazilian Ministry of the Environment: Brasilia, Brazil, 2005. [Google Scholar]
- Patterson, R.A. A Resident’s Role in Minimising Nitrogen, Phosphorus and Salt in Domestic Wastewater. In Proceedings of the Tenth National Symposium on Individual and Small Community Sewage Systems Proceedings, Sacramento, CA, USA, 21–24 March 2004; pp. 740–749. [Google Scholar]
- De Jonckheere, J.F.; Pernin, P.; Scaglia, M.; Michel, R. A comparative study of 14 strains of Naegleria australiensis demonstrates the existence of a highly virulent subspecies: N. australiensis italica n. spp. J. Protozool. 1984, 31, 324–331. [Google Scholar] [CrossRef]
- Scaglia, M.; Strosselli, M.; Grazioli, V.; Gatti, S.; Bernuzzi, A.M.; de Jonckheere, J.F. Isolation and identification of pathogenic Naegleria australiensis (Amoebida, Vahlkampfidae) from a spa in northern Italy. Appl. Environ. Microbiol. 1983, 46, 1282–1285. [Google Scholar] [CrossRef] [Green Version]
- Dyková, I.; Kyselová, I.; Pecková, H.; Oborník, M.; Lukes, J. Identity of Naegleria strains isolated from organs of freshwater fishes. Dis. Aquat. Org. 2001, 46, 115–121. [Google Scholar] [CrossRef]
- Visvesvara, G.S.; Moura, H.; Schuster, F.L. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol. Med. Microbiol. 2007, 50, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Schuster, F.L.; Visvesvara, G.S. Free-living amoebae as opportunistic and non-opportunistic pathogens of humans and animals. Int. J. Parasitol. 2004, 34, 1001–1027. [Google Scholar] [CrossRef] [PubMed]
- Petit, F.; Vilchez, V.; Torres, G.; Molina, O.; Dorfman, S.; Mora, E.; Cardozo, J. Meningoencefalitis amebiana primaria: comunicacion de dos nuevos casos Venezolanos. Arq. de Neuro-Psiquiatria 2006, 64, 1043–1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz, V.; Reyes, H.; Toche, P.; Carcamo, C.; Gottlieb, B. Aislamiento de amebas de vida libre en piscinas públicas de Santiago de Chile. Parasitología latinoamericana 2003, 58, 106–111. [Google Scholar] [CrossRef]
- Kao, P.-M.; Hsu, B.-M.; Hsu, T.-K.; Chiu, Y.-C.; Chang, C.-L.; Ji, W.-T.; Huang, S.-W.; Fan, C.-W. Application of TaqMan qPCR for the detection and monitoring of Naegleria species in reservoirs used as a source for drinking water. Parasitol. Res. 2014, 113, 3765–3771. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Batlle, M.; Hernández-Piñero, I.; Rizo-Liendo, A.; López-Arencibia, A.; Sifaoui, I.; Bethencourt-Estrella, C.J.; Chiboub, O.; Valladares, B.; Piñero, J.E.; Lorenzo-Morales, J. Isolation and molecular identification of free-living amoebae from dishcloths in Tenerife, Canary Islands, Spain. Parasitol. Res. 2019, 118, 927–933. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]





| Sample Sites | Culture Temperature | 1-Isolates from NNA Cultures | 2-Isolates Directly from Water Samples | ||
|---|---|---|---|---|---|
| Code | BLASTn/Accession 1 | Code | BLASTn/Accession 1 | ||
| A | 26 ℃ | N2.G2 | N. philippinensis/LC191904.1 | N1 | N. canariensis/FJ475124.1 |
| 37 ℃ | N1 | N. canariensis/FJ475124.1 | |||
| B | 26 ℃ | N1 | N. canariensis/FJ475124.1 | N2.G1 N1 | N. philippinensis; N. canariensis/FJ475124.1 |
| C | 26 ℃ | N1 | N. canariensis/FJ475124.1 | N4.G1 N3.G1 N3.G2 | N. australiensis/AB128053.1; N. dobsoni/KU380484.1 N. dobsoni/KU380484.1 |
| 37 ℃ | N1 | N. canariensis/FJ475124.1 | |||
| D | 26 ℃ | N4.G2 | N. australiensis/AB128052.1 | N2.G1 N5.G1 | N. philippinensis/AY033618.1; N. gruberi/MG699123.1 |
| 37 ℃ | N2.G2 | N. philippinensis/LC191904.1 | |||
| H1 | Hartmannella/HE617186.1 | ||||
| 44 ℃ | N4.G1 | N. australiensis/AB128053.1 | |||
| E | 26 ℃ | N5.G2 | N. gruberi/MG699123.1 | N5.G2 N1 | N. gruberi/MG699123.1; N. canariensis/FJ475124.1 |
| 37 ℃ | N1 | N. canariensis/FJ475124.1 | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bellini, N.K.; Fonseca, A.L.M.d.; Reyes-Batlle, M.; Lorenzo-Morales, J.; Rocha, O.; Thiemann, O.H. Isolation of Naegleria spp. from a Brazilian Water Source. Pathogens 2020, 9, 90. https://doi.org/10.3390/pathogens9020090
Bellini NK, Fonseca ALMd, Reyes-Batlle M, Lorenzo-Morales J, Rocha O, Thiemann OH. Isolation of Naegleria spp. from a Brazilian Water Source. Pathogens. 2020; 9(2):90. https://doi.org/10.3390/pathogens9020090
Chicago/Turabian StyleBellini, Natália Karla, Ana Letícia Moreira da Fonseca, María Reyes-Batlle, Jacob Lorenzo-Morales, Odete Rocha, and Otavio Henrique Thiemann. 2020. "Isolation of Naegleria spp. from a Brazilian Water Source" Pathogens 9, no. 2: 90. https://doi.org/10.3390/pathogens9020090
APA StyleBellini, N. K., Fonseca, A. L. M. d., Reyes-Batlle, M., Lorenzo-Morales, J., Rocha, O., & Thiemann, O. H. (2020). Isolation of Naegleria spp. from a Brazilian Water Source. Pathogens, 9(2), 90. https://doi.org/10.3390/pathogens9020090