The Laboratory Diagnosis of Neisseria gonorrhoeae: Current Testing and Future Demands
Abstract
:1. Introduction
- Improvements of Ng testing resulted in increased detection rates that may have influenced epidemiologic data (i.e., higher detection rates do not necessarily indicate an increase in transmitted infections but may just reflect more sensitive and more frequent testing). For instance, introduction of NAATs in routine diagnostic testing have shown pharyngeal and rectal infection to be much more prevalent than previously assumed [14].
- Since rectal and pharyngeal infections, as well as cervical infections in women, are frequently asymptomatic and will be missed by symptom-based examinations, laboratory testing should consider inclusion of both urogenital, anorectal, and pharyngeal samples, depending on sexual behavior, to identify infected individuals with higher sensitivity [5,15,16].
- NAAT-based treatment monitoring has improved identification of treatment failures that particularly relate to pharyngeal infections [17]. Considering the presence of non-gonococcal Neisseria species at the pharyngeal mucosa that may transfer resistance to Ng [18,19], the pharynx has been suggested an important site for resistance development. Currently, the frequency and impact of genetic exchange in the pharynx is not known exactly but is important to be clarified, as it would strongly support pharyngeal screening and clearance of pharyngeal infections to be essential.
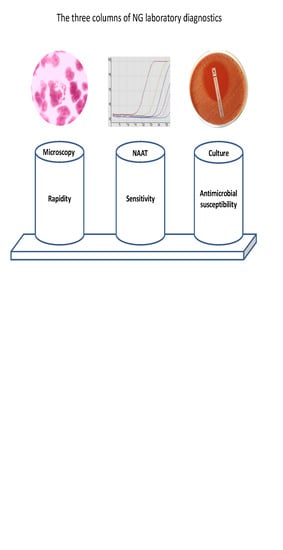
2. Microscopy
3. Culture
4. Antimicrobial Susceptibility Testing
5. NAATs
6. Rapid Tests and Point-of-Care Tests (POCT)
7. Future Demands
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rowley, J.; Vander Hoorn, S.; Korenromp, E.; Low, N.; Unemo, M.; Abu-Raddad, L.J.; Chico, R. Chlamydia, gonorrhoea, trichomoniasis and syphilis: Global prevalence and incidence estimates, 2016. Bull World Heal. Organ. 2019, 97, 548–562. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Gonorrhoea—Annual Epidemiological Report for 2017; European Centre for Disease Prevention and Control: Stockholm, Sweden, 2019. [Google Scholar]
- Department of Health and Human Services. Sexually Transmitted Disease Surveillance 2018; Centers for Disease Control and Prevention: Atlanta, GE, USA, 2019. [Google Scholar] [CrossRef]
- Fifer, H.; Saunders, J.; Soni, S.; Sadiq, T.; FitzGerald, M. National Guideline for the Management of Infection with Neisseria gonorrhoeae. British Association for Sexual Health and HIV Web Site, 2019. Available online: https://www.bashhguidelines.org/media/1208/gc-2019.pdf (accessed on 30 January 2020).
- Dudareva-Vizule, S.; Haar, K.; Sailer, A.; Wisplinghoff, H.; Wisplinghoff, F.; Marcus, U.; PARIS Study Group. Prevalence of pharyngeal and rectal Chlamydia trachomatis and Neisseria gonorrhoeae infections among men who have sex with men in Germany. Sex Transm. Infect. 2014, 90, 46–51. [Google Scholar] [CrossRef]
- Buder, S.; Dudareva, S.; Jansen, K.; Loenenbach, A.; Nikisins, S.; Sailer, A.; Guhl, E.; Kohl, P.K.; Bremer, V.; GORENET Study Group. Antimicrobial resistance of Neisseria gonorrhoeae in Germany: Low levels of cephalosporin resistance, but high azithromycin resistance. BMC Infect. Dis. 2018, 18, 44. [Google Scholar] [CrossRef] [Green Version]
- McConaghy, J.R.; Panchal, B. Epididymitis: An Overview. Am. Fam. Physician 2016, 94, 723–726. [Google Scholar]
- Reekie, J.; Donovan, B.; Guy, R.; Hocking, J.S.; Kaldor, J.M.; Mak, D.; Preen, D.; Ward, J.; Liu, B. Risk of Pelvic Inflammatory Disease in Relation to Chlamydia and Gonorrhea Testing, Repeat Testing, and Positivity: A Population-Based Cohort Study. Clin. Infect. Dis. 2018, 66, 437–443. [Google Scholar] [CrossRef]
- Brunham, R.C.; Gottlieb, S.L.; Paavonen, J. Pelvic inflammatory disease. N. Engl. J. Med. 2015, 372, 2039–2048. [Google Scholar] [CrossRef]
- Belkacem, A.; Caumes, E.; Ouanich, J.; Jarlier, V.; Dellion, S.; Cazenave, B.; Goursaud, R.; Lacassin, F.; Breuil, J.; Patey, O.; et al. Changing patterns of disseminated gonococcal infection in France: Cross-sectional data 2009–2011. Sex Transm. Infect. 2013, 89, 613–615. [Google Scholar] [CrossRef]
- Heumann, C.L.; Quilter, L.A.; Eastment, M.C.; Heffron, R.; Hawes, S.E. Adverse Birth Outcomes and Maternal Neisseria gonorrhoeae Infection: A Population-Based Cohort Study in Washington State. Sex Transm. Dis. 2017, 44, 266–271. [Google Scholar] [CrossRef] [Green Version]
- Alexander, E.R. Gonorrhea in the newborn. Ann. N. Y. Acad. Sci. 1988, 549, 180–186. [Google Scholar] [CrossRef]
- Papp, J.R.; Schachter, J.; Gaydos, C.A.; van der Pol, B. Recommendations for the laboratory-based detection of Chlamydia trachomatis and Neisseria gonorrhoeae—2014. MMWR Recomm. Rep. 2014, 63, 1–19. [Google Scholar]
- Cornelisse, V.J.; Chow, E.P.; Huffam, S.; Fairley, C.K.; Bissessor, M.; De Petra, V.; Howden, B.P.; Denham, I.; Bradshaw, C.S.; Williamson, D.; et al. Increased Detection of Pharyngeal and Rectal Gonorrhea in Men Who Have Sex With Men After Transition From Culture To Nucleic Acid Amplification Testing. Sex Transm. Dis. 2017, 44, 114–117. [Google Scholar] [CrossRef]
- Kent, C.K.; Chaw, J.K.; Wong, W.; Liska, S.; Gibson, S.; Hubbard, G.; Klausner, J.D. Prevalence of rectal, urethral, and pharyngeal Chlamydia and gonorrhea detected in 2 clinical settings among men who have sex with men: San Francisco, California, 2003. Clin. Infect. Dis. 2005, 41, 67–74. [Google Scholar] [CrossRef]
- Weston, E.J.; Kirkcaldy, R.D.; Stenger, M.; Llata, E.; Hoots, B.; Torrone, E.A. Narrative Review: Assessment of Neisseria gonorrhoeae Infections Among Men Who Have Sex With Men in National and Sentinel Surveillance Systems in the United States. Sex Transm. Dis. 2018, 45, 243–249. [Google Scholar] [CrossRef]
- Allen, V.G.; Mitterni, L.; Seah, C.; Rebbapragada, A.; Martin, I.E.; Lee, C.; Siebert, H.; Towns, L.; Melano, R.G.; Low, D.E. Neisseria gonorrhoeae treatment failure and susceptibility to cefixime in Toronto, Canada. JAMA 2013, 309, 163–170. [Google Scholar] [CrossRef] [Green Version]
- Spratt, B.G.; Bowler, L.D.; Zhang, Q.Y.; Zhou, J.; Smith, J.M. Role of interspecies transfer of chromosomal genes in the evolution of penicillin resistance in pathogenic and commensal Neisseria species. J. Mol. Evol. 1992, 34, 115–125. [Google Scholar] [CrossRef]
- Wadsworth, C.B.; Arnold, B.J.; Sater, M.R.A.; Grad, Y.H. Azithromycin Resistance through Interspecific Acquisition of an Epistasis-Dependent Efflux Pump Component and Transcriptional Regulator in Neisseria gonorrhoeae. MBio 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Unemo, M.; Seifert, H.S.; Hook, E.W., III; Hawkes, S.; Ndowa, F.; Dillon, J.R. Gonorrhoea. Nat. Rev. Dis. Primers 2019, 5, 79. [Google Scholar] [CrossRef]
- Thorley, N.; Radcliffe, K. The performance and clinical utility of cervical microscopy for the diagnosis of gonorrhoea in women in the era of the NAAT. Int. J. STD AIDS 2015, 26, 656–660. [Google Scholar] [CrossRef]
- Mensforth, S.; Thorley, N.; Radcliffe, K. Auditing the use and assessing the clinical utility of microscopy as a point-of-care test for Neisseria gonorrhoeae in a Sexual Health clinic. Int. J. STD AIDS 2018, 29, 157–163. [Google Scholar] [CrossRef]
- Unemo, M.; Ison, C. Gonorrhea. In Laboratory Diagnosis of Sexually Transmitted Infections, Including Human Immunodeficiency Virus; Unemo, M., Ballard, R., Ison, C., Lewis, D., Ndowna, F., Peeling, R., Eds.; WHO: Geneva, Switzerland, 2013; Available online: https://www.who.int/reproductivehealth/publications/rtis/9789241505840/en/ (accessed on 30 January 2020).
- Bignell, C.; Ison, C.A.; Jungmann, E. Gonorrhoea. Sex Transm. Infect. 2006, 82 (Suppl. 4), iv6–iv9. [Google Scholar] [CrossRef]
- Ota, K.V.; Tamari, I.E.; Smieja, M.; Jamieson, F.; Jones, K.E.; Towns, L.; Juzkiw, J.; Richardson, S.E. Detection of Neisseria gonorrhoeae and Chlamydia trachomatis in pharyngeal and rectal specimens using the BD Probetec ET system, the Gen-Probe Aptima Combo 2 assay and culture. Sex Transm. Infect. 2009, 85, 182–186. [Google Scholar] [CrossRef]
- Alexander, S. The challenges of detecting gonorrhoea and Chlamydia in rectal and pharyngeal sites: Could we, should we, be doing more? Sex Transm. Infect. 2009, 85, 159–160. [Google Scholar] [CrossRef]
- Visser, M.; van Westreenen, M.; van Bergen, J.; van Benthem, B.H.B. Low gonorrhoea antimicrobial resistance and culture positivity rates in general practice: A pilot study. Sex Transm. Infect. 2019. [Google Scholar] [CrossRef] [Green Version]
- Alexander, S.; Ison, C. Evaluation of commercial kits for the identification of Neisseria gonorrhoeae. J. Med. Microbiol. 2005, 54, 827–831. [Google Scholar] [CrossRef]
- Dillon, J.R.; Carballo, M.; Pauzé, M. Evaluation of eight methods for identification of pathogenic Neisseria species: Neisseria-Kwik, RIM-N, Gonobio-Test, Minitek, Gonochek II, GonoGen, Phadebact Monoclonal GC OMNI Test, and Syva MicroTrak Test. J. Clin. Microbiol. 1988, 26, 493–497. [Google Scholar]
- Carannante, A.; De Carolis, E.; Vacca, P.; Vella, A.; Vocale, C.; De Francesco, M.A.; Cusini, M.; Del Re, S.; Dal Conte, I.; Cristaudo, A.; et al. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) for identification and clustering of Neisseria gonorrhoeae. BMC Microbiol. 2015, 15, 142. [Google Scholar] [CrossRef] [Green Version]
- Buchanan, R.; Ball, D.; Dolphin, H.; Dave, J. Matrix-assisted laser desorption-ionization time-of-flight mass spectrometry for the identification of Neisseria gonorrhoeae. Clin. Microbiol. Infect. 2016, 22, 815. [Google Scholar] [CrossRef] [Green Version]
- Schweitzer, V.A.; van Dam, A.P.; Hananta, I.P.; Schuurman, R.; Kusters, J.G.; Rentenaar, R.J. Identification of Neisseria gonorrhoeae by the Bruker Biotyper Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry System Is Improved by a Database Extension. J. Clin. Microbiol. 2016, 54, 1130–1132. [Google Scholar] [CrossRef] [Green Version]
- Morel, F.; Jacquier, H.; Desroches, M.; Fihman, V.; Kumanski, S.; Cambau, E.; Decousser, J.W.; Berçot, B. Use of Andromas and Bruker MALDI-TOF MS in the identification of Neisseria. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 2273–2277. [Google Scholar] [CrossRef]
- Unemo, M.; Fasth, O.; Fredlund, H.; Limnios, A.; Tapsall, J. Phenotypic and genetic characterization of the 2008 WHO Neisseria gonorrhoeae reference strain panel intended for global quality assurance and quality control of gonococcal antimicrobial resistance surveillance for public health purposes. J. Antimicrob. Chemother. 2009, 63, 1142–1151. [Google Scholar] [CrossRef]
- Unemo, M.; Golparian, D.; Sánchez-Busó, L.; Grad, Y.; Jacobsson, S.; Ohnishi, M.; Lahra, M.M.; Limnios, A.; Sikora, A.E.; Wi, T.; et al. The novel 2016 WHO Neisseria gonorrhoeae reference strains for global quality assurance of laboratory investigations: Phenotypic, genetic and reference genome characterization. J. Antimicrob. Chemother. 2016, 71, 3096–3108. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Taylor, T.H., Jr.; Pettus, K.; Johnson, S.; Papp, J.R.; Trees, D. Comparing the disk-diffusion and agar dilution tests for Neisseria gonorrhoeae antimicrobial susceptibility testing. Antimicrob. Resist. Infect. Control 2016, 5, 46. [Google Scholar] [CrossRef] [Green Version]
- Singh, V.; Bala, M.; Kakran, M.; Ramesh, V. Comparative assessment of CDS, CLSI disc diffusion and Etest techniques for antimicrobial susceptibility testing of Neisseria gonorrhoeae: A 6-year study. BMJ Open 2012, 2. [Google Scholar] [CrossRef] [Green Version]
- Mal, P.B.; Jabeen, K.; Farooqi, J.; Unemo, M.; Khan, E. Antimicrobial susceptibility testing of Neisseria gonorrhoeae isolates in Pakistan by Etest compared to Calibrated Dichotomous Sensitivity and Clinical Laboratory Standards Institute disc diffusion techniques. BMC Microbiol. 2016, 16, 236. [Google Scholar] [CrossRef] [Green Version]
- Enriquez, R.P.; Goire, N.; Kundu, R.; Gatus, B.J.; Lahra, M.M. A comparison of agar dilution with the Calibrated Dichotomous Sensitivity (CDS) and Etest methods for determining the minimum inhibitory concentration of ceftriaxone against Neisseria gonorrhoeae. Diagn. Microbiol. Infect. Dis. 2016, 86, 40–43. [Google Scholar] [CrossRef]
- Foerster, S.; Desilvestro, V.; Hathaway, L.J.; Althaus, C.L.; Unemo, M. A new rapid resazurin-based microdilution assay for antimicrobial susceptibility testing of Neisseria gonorrhoeae. J. Antimicrob. Chemother. 2017, 72, 1961–1968. [Google Scholar] [CrossRef]
- Liu, H.; Taylor, T.H., Jr.; Pettus, K.; Trees, D. Assessment of Etest as an alternative to agar dilution for antimicrobial susceptibility testing of Neisseria gonorrhoeae. J. Clin. Microbiol. 2014, 52, 1435–1440. [Google Scholar] [CrossRef] [Green Version]
- Papp, J.R.; Rowlinson, M.C.; O’Connor, N.P.; Wholehan, J.; Razeq, J.H.; Glennen, A.; Ware, D.; Iwen, P.C.; Lee, L.V.; Hagan, C. Accuracy and reproducibility of the Etest to detect drug-resistant Neisseria gonorrhoeae to contemporary treatment. J. Med. Microbiol. 2018, 67, 68–73. [Google Scholar] [CrossRef] [Green Version]
- Jönsson, A.; Jacobsson, S.; Foerster, S.; Cole, M.J.; Unemo, M. Performance characteristics of newer MIC gradient strip tests compared with the Etest for antimicrobial susceptibility testing of Neisseria gonorrhoeae. APMIS 2018, 126, 822–827. [Google Scholar] [CrossRef]
- McAuliffe, G.N.; Smith, M.; Cooper, G.; Forster, R.F.; Roberts, S.A. Variability in Azithromycin Susceptibility Results for Neisseria gonorrhoeae Obtained Using Gradient MIC Strip and Agar Dilution Techniques. J. Clin. Microbiol. 2019, 57, e01353–19. [Google Scholar] [CrossRef]
- Tapsall, J.W.; Phillips, E.A.; Morris, L.M. Chromosomally mediated intrinsic resistance to penicillin in penicillinase producing strains of Neisseria gonorrhoeae isolated in Sydney: Guide to treatment with Augmentin. Genitourin. Med. 1987, 63, 305–308. [Google Scholar] [CrossRef] [Green Version]
- Khazaei, T.; Barlow, J.T.; Schoepp, N.G.; Ismagilov, R.F. RNA markers enable phenotypic test of antibiotic susceptibility in Neisseria gonorrhoeae after 10 minutes of ciprofloxacin exposure. Sci. Rep. 2018, 8, 11606. [Google Scholar] [CrossRef] [Green Version]
- Wadsworth, C.B.; Sater, M.R.A.; Bhattacharyya, R.P.; Grad, Y.H. Impact of Species Diversity on the Design of RNA-Based Diagnostics for Antibiotic Resistance in Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.H.; Qin, X.L.; Yang, J.Y.; Liao, Y.W.; Wu, X.Z.; Zheng, H.P. Identification and expression analysis of ceftriaxone resistance-related genes in Neisseria gonorrhoeae integrating RNA-Seq data and qRT-PCR validation. J. Glob. Antimicrob. Resist. 2019, 16, 202–209. [Google Scholar] [CrossRef]
- Martin, D.H.; Cammarata, C.; van der Pol, B.; Jones, R.B.; Quinn, T.C.; Gaydos, C.A.; Crotchfelt, K.; Schachter, J.; Moncada, J.; Jungkind, D.; et al. Multicenter evaluation of AMPLICOR and automated cobas AMPLICOR CTT/NG tests for Neisseria gonorrhoeae. J. Clin. Microbiol. 2000, 38, 3544–3549. [Google Scholar]
- Cook, R.L.; Hutchison, S.L.; Ostergaard, L.; Braithwaite, R.S.; Ness, R.B. Systematic review: Noninvasive testing for Chlamydia trachomatis and Neisseria gonorrhoeae. Ann. Intern. Med. 2005, 142, 914–925. [Google Scholar] [CrossRef]
- Van Dyck, E.; Ieven, M.; Pattyn, S.; Van Damme, L.; Laga, M. Detection of Chlamydia trachomatis and Neisseria gonorrhoeae by enzyme immunoassay, culture, and three nucleic acid amplification tests. J. Clin. Microbiol. 2001, 39, 1751–1756. [Google Scholar] [CrossRef] [Green Version]
- Serra-Pladevall, J.; Caballero, E.; Roig, G.; Juvé, R.; Barbera, M.J.; Andreu, A. Comparison between conventional culture and NAATs for the microbiological diagnosis in gonococcal infection. Diagn. Microbiol. Infect. Dis. 2015, 83, 341–343. [Google Scholar] [CrossRef]
- Bromhead, C.; Miller, A.; Jones, M.; Whiley, D. Comparison of the cobas 4800 CT/NG test with culture for detecting Neisseria gonorrhoeae in genital and nongenital specimens in a low-prevalence population in New Zealand. J. Clin. Microbiol. 2013, 51, 1505–1509. [Google Scholar] [CrossRef] [Green Version]
- Van der Pol, B.; Hook, E.W., III; Williams, J.A.; Smith, B.; Taylor, S.N. Performance of the BD CTQx and GCQx Amplified Assays on the BD Viper LT Compared With the BD Viper XTR System. Sex Transm. Dis. 2015, 42, 521–523. [Google Scholar] [CrossRef]
- Cheng, A.; Kirby, J.E. Evaluation of the Hologic gen-probe PANTHER, APTIMA Combo 2 assay in a tertiary care teaching hospital. Am. J. Clin. Pathol. 2014, 141, 397–403. [Google Scholar] [CrossRef] [Green Version]
- Marlowe, E.M.; Hardy, D.; Krevolin, M.; Gohl, P.; Bertram, A.; Arcenas, R.; Seiverth, B.; Schneider, T.; Liesenfeld, O. High-throughput testing of urogenital and extragenital specimens for detection of Chlamydia trachomatis and Neisseria gonorrhoeae with Cobas CT/NG. Eur. J. Microbiol. Immunol. Bp. 2017, 7, 176–186. [Google Scholar] [CrossRef] [Green Version]
- Van der Pol, B.; Williams, J.A.; Fuller, D.; Taylor, S.N.; Hook, E.W., III. Combined Testing for Chlamydia, gonorrhea, and Trichomonas by use of the BD Max CT/GC/TV Assay with genitourinary specimen types. J. Clin. Microbiol. 2017, 55, 155–164. [Google Scholar] [CrossRef] [Green Version]
- Whiley, D.M.; Limnios, A.; Moon, N.J.; Gehrig, N.; Goire, N.; Hogan, T.; Lam, A.; Jacob, K.; Lambert, S.B.; Nissen, M.D.; et al. False-negative results using Neisseria gonorrhoeae porA pseudogene PCR—A clinical gonococcal isolate with an N. meningitidis porA sequence, Australia, March 2011. Eurosurveillance 2011, 16, 19874. [Google Scholar]
- Ison, C.A.; Golparian, D.; Saunders, P.; Chisholm, S.; Unemo, M. Evolution of Neisseria gonorrhoeae is a continuing challenge for molecular detection of gonorrhoea: False negative gonococcal porA mutants are spreading internationally. Sex Transm. Infect. 2013, 89, 197–201. [Google Scholar] [CrossRef] [Green Version]
- Upton, A.; Bromhead, C.; Whiley, D.M. Neisseria gonorrhoeae false-positive result obtained from a pharyngeal swab by using the Roche cobas 4800 CT/NG assay in New Zealand in 2012. J. Clin. Microbiol. 2013, 51, 1609–1610. [Google Scholar] [CrossRef] [Green Version]
- Frosch, M.; Meyer, T.F. Transformation-mediated exchange of virulence determinants by co-cultivation of pathogenic Neisseriae. FEMS Microbiol. Lett. 1992, 100, 345–349. [Google Scholar] [CrossRef]
- Li, J.; Jang, D.; Gilchrist, J.; Smieja, M.; Ewert, R.; MacRitchie, C.; Chernesky, M. Comparison of flocked and aptima swabs and two specimen transport media in the aptima combo 2 assay. J. Clin. Microbiol. 2014, 52, 3808–3809. [Google Scholar] [CrossRef] [Green Version]
- Wind, C.M.; de Vries, H.J.; Schim van der Loeff, M.F.; Unemo, M.; van Dam, A.P. Successful Combination of Nucleic Acid Amplification Test Diagnostics and Targeted Deferred Neisseria gonorrhoeae Culture. J. Clin. Microbiol. 2015, 53, 1884–1990. [Google Scholar] [CrossRef] [Green Version]
- Schachter, J.; Chernesky, M.A.; Willis, D.E.; Fine, P.M.; Martin, D.H.; Fuller, D.; Jordan, J.A.; Janda, W.; Hook, E.W., III. Vaginal swabs are the specimens of choice when screening for Chlamydia trachomatis and Neisseria gonorrhoeae: Results from a multicenter evaluation of the APTIMA assays for both infections. Sex Transm. Dis. 2005, 32, 725–728. [Google Scholar] [CrossRef]
- Stewart, C.M.; Schoeman, S.A.; Booth, R.A.; Smith, S.D.; Wilcox, M.H.; Wilson, J.D. Assessment of self taken swabs versus clinician taken swab cultures for diagnosing gonorrhoea in women: Single centre, diagnostic accuracy study. BMJ Clin. Res. Ed. 2012, 345, e8107. [Google Scholar] [CrossRef] [Green Version]
- Dize, L.; Agreda, P.; Quinn, N.; Barnes, M.R.; Hsieh, Y.H.; Gaydos, C.A. Comparison of self-obtained penile-meatal swabs to urine for the detection of C. trachomatis, N. gonorrhoeae and T. vaginalis. Sex Transm. Infect. 2013, 89, 305–307. [Google Scholar] [CrossRef] [Green Version]
- AWMF. Leitlinie: Sexuell Übertragbare Infektionen (STI)—Beratung, Diagnostik, Therapie, 059-006. 2019. Available online: https://www.awmf.org/uploads/tx_szleitlinien/059-006l_S2k_Sexuell-uebertragbare-Infektionen-Beratung-Diagnostik-Therapie-STI_2019-09.pdf (accessed on 30 January 2020).
- Page-Shafer, K.; Graves, A.; Kent, C.; Balls, J.E.; Zapitz, V.M.; Klausner, J.D. Increased sensitivity of DNA amplification testing for the detection of pharyngeal gonorrhea in men who have sex with men. Clin. Infect. Dis. 2002, 34, 173–176. [Google Scholar] [CrossRef]
- Schachter, J.; Moncada, J.; Liska, S.; Shayevich, C.; Klausner, J.D. Nucleic acid amplification tests in the diagnosis of chlamydial and gonococcal infections of the oropharynx and rectum in men who have sex with men. Sex Transm. Dis. 2008, 35, 637–642. [Google Scholar] [CrossRef] [Green Version]
- Hughes, G.; Ison, C.; Field, N. Guidance for the Detection of Gonorrhoea in England; Public Health England: London, UK, 2014; Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/769003/170215_Gonorrhoea_testing_guidance_REVISED__2_.pdf. (accessed on 30 January 2020).
- Benn, P.D.; Rooney, G.; Carder, C.; Brown, M.; Stevenson, S.R.; Copas, A.; Robinson, A.J.; Ridgway, G.L. Chlamydia trachomatis and Neisseria gonorrhoeae infection and the sexual behaviour of men who have sex with men. Sex Transm. Infect. 2007, 83, 106–112. [Google Scholar] [CrossRef] [Green Version]
- Fifer, H.; Natarajan, U.; Jones, L.; Alexander, S.; Hughes, G.; Golparian, D.; Unemo, M. Failure of Dual Antimicrobial Therapy in Treatment of Gonorrhea. N. Engl. J. Med. 2016, 374, 2504–2506. [Google Scholar] [CrossRef]
- Palmer, H.M.; Mallinson, H.; Wood, R.L.; Herring, A.J. Evaluation of the specificities of five DNA amplification methods for the detection of Neisseria gonorrhoeae. J. Clin. Microbiol. 2003, 41, 835–837. [Google Scholar] [CrossRef] [Green Version]
- Field, N.; Clifton, S.; Alexander, S.; Ison, C.A.; Hughes, G.; Beddows, S.; Tanton, C.; Soldan, K.; Coelho da Silva, F.; Mercer, C.H.; et al. Confirmatory assays are essential when using molecular testing for Neisseria gonorrhoeae in low-prevalence settings: Insights from the third National Survey of Sexual Attitudes and Lifestyles (Natsal-3). Sex Transm. Infect. 2015, 91, 338–341. [Google Scholar] [CrossRef] [Green Version]
- Chow, E.P.; Fehler, G.; Read, T.R.; Tabrizi, S.N.; Hocking, J.S.; Denham, I.; Bradshaw, C.S.; Chen, M.Y.; Fairley, C.K. Gonorrhoea notifications and nucleic acid amplification testing in a very low-prevalence Australian female population. Med. J. Aust. 2015, 202, 321–323. [Google Scholar] [CrossRef] [Green Version]
- Tabrizi, S.N.; Unemo, M.; Golparian, D.; Twin, J.; Limnios, A.E.; Lahra, M.; Guy, R.; TTANGO Investigators. Analytical evaluation of GeneXpert CT/NG, the first genetic point-of-care assay for simultaneous detection of Neisseria gonorrhoeae and Chlamydia trachomatis. J. Clin. Microbiol. 2013, 51, 1945–1947. [Google Scholar] [CrossRef] [Green Version]
- Perry, M.D.; Jones, R.N.; Corden, S.A. Is confirmatory testing of Roche cobas 4800 CT/NG test Neisseria gonorrhoeae positive samples required? Comparison of the Roche cobas 4800 CT/NG test with an opa/pap duplex assay for the detection of N. gonorrhoeae. Sex Transm. Infect. 2014, 90, 303–308. [Google Scholar] [CrossRef]
- Ursi, D.; Crucitti, T.; Smet, H.; Ieven, M. Evaluation of the Bio-Rad Dx CT/NG/MG® assay for simultaneous detection of Chlamydia trachomatis, Neisseria gonorrhoeae and Mycoplasma genitalium in urine. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1159–1163. [Google Scholar] [CrossRef]
- Rumyantseva, T.; Golparian, D.; Nilsson, C.S.; Johansson, E.; Falk, M.; Fredlund, H.; Van Dam, A.; Guschin, A.; Unemo, M. Evaluation of the new AmpliSens multiplex real-time PCR assay for simultaneous detection of Neisseria gonorrhoeae, Chlamydia trachomatis, Mycoplasma genitalium and Trichomonas vaginalis. APMIS 2015, 123, 879–886. [Google Scholar] [CrossRef]
- Kriesel, J.D.; Bhatia, A.S.; Barrus, C.; Vaughn, M.; Gardner, J.; Crisp, R.J. Multiplex PCR testing for nine different sexually transmitted infections. Int. J. STD. AIDS 2016, 27, 1275–1282. [Google Scholar] [CrossRef]
- Choe, H.S.; Lee, D.S.; Lee, S.J.; Hong, S.H.; Park, D.C.; Lee, M.K.; Kim, T.H.; Cho, Y.H. Performance of Anyplex™ II multiplex real-time PCR for the diagnosis of seven sexually transmitted infections: Comparison with currently available methods. Int. J. Infect. Dis. 2013, 17, e1134–40. [Google Scholar] [CrossRef] [Green Version]
- Berçot, B.; Amarsy, R.; Goubard, A.; Aparicio, C.; Loeung, H.U.; Segouin, C.; Gueret, H.; Jacquier, F.; Meunier, F.; Mougari, E.; et al. Assessment of coinfection of sexually transmitted pathogen microbes by use of the anyplex II STI-7 molecular kit. J. Clin. Microbiol. 2015, 53, 991–993. [Google Scholar] [CrossRef] [Green Version]
- Bignell, C.; Unemo, M. European STI Guidelines Editorial Board. 2012 European guideline on the diagnosis and treatment of gonorrhoea in adults. Int. J. STD. AIDS 2013, 24, 85–92. [Google Scholar] [CrossRef]
- Guy, R.J.; Causer, L.M.; Klausner, J.D.; Unemo, M.; Toskin, I.; Azzini, A.M.; Peeling, R.W. Performance and operational characteristics of point-of-care tests for the diagnosis of urogenital gonococcal infections. Sex Transm. Infect. 2017, 93, S16–S21. [Google Scholar] [CrossRef]
- Abbai, N.S.; Moodley, P.; Reddy, T.; Zondi, T.G.; Rambaran, S.; Naidoo, K.; Ramjee, G. Clinical evaluation of the OneStep Gonorrhea RapiCard InstaTest for detection of Neisseria gonorrhoeae in symptomatic patients from KwaZulu-Natal, South Africa. J. Clin. Microbiol. 2015, 53, 1348–1350. [Google Scholar] [CrossRef] [Green Version]
- Benzaken, A.S.; Galban, E.G.; Antunes, W.; Dutra, J.C.; Peeling, R.W.; Mabey, D.; Salama, A. Diagnosis of gonococcal infection in high risk women using a rapid test. Sex Transm. Infect. 2006, 82 (Suppl. 5), v26–v28. [Google Scholar] [CrossRef]
- Suzuki, K.; Matsumoto, T.; Murakami, H.; Tateda, K.; Ishii, N.; Yamaguchi, K. Evaluation of a rapid antigen detection test for Neisseria gonorrhoeae in urine sediment for diagnosis of gonococcal urethritis in males. J. Infect. Chemother. 2004, 10, 208–211. [Google Scholar] [CrossRef]
- Nuñez-Forero, L.; Moyano-Ariza, L.; Gaitán-Duarte, H.; Ángel-Müller, E.; Ruiz-Parra, A.; González, P.; Rodríguez, A.; Tolosa, J.E. Diagnostic accuracy of rapid tests for sexually transmitted infections in symptomatic women. Sex Transm. Infect. 2016, 92, 24–28. [Google Scholar] [CrossRef]
- Samarawickrama, A.; Cheserem, E.; Graver, M.; Wade, J.; Alexander, S.; Ison, C. Pilot study of use of the BioStar Optical ImmunoAssay GC point-of-care test for diagnosing gonorrhoea in men attending a genitourinary medicine clinic. J. Med. Microbiol. 2014, 63, 1111–1112. [Google Scholar] [CrossRef] [Green Version]
- Peeling, R.W.; Holmes, K.K.; Mabey, D.; Ronald, A. Rapid tests for sexually transmitted infections (STIs): The way forward. Sex Transm. Infect. 2006, 82, v1–v6. [Google Scholar] [CrossRef]
- Gaydos, C.A.; van der Pol, B.; Jett-Goheen, M.; Barnes, M.; Quinn, N.; Clark, C.; Daniel, G.E.; Dixon, P.B.; Hook, E.W., III; CT/NG Study Group. Performance of the Cepheid CT/ NG Xpert Rapid PCR Test for Detection of Chlamydia trachomatis and Neisseria gonorrhoeae. J. Clin. Microbiol. 2013, 51, 1666–1672. [Google Scholar] [CrossRef] [Green Version]
- Gaydos, C.A. Review of use of a new rapid real-time PCR, the Cepheid GeneXpert® (Xpert) CT/NG assay, for Chlamydia trachomatis and Neisseria gonorrhoeae: Results for patients while in a clinical setting. Expert. Rev. Mol. Diagn. 2014, 14, 135–137. [Google Scholar] [CrossRef] [Green Version]
- Garrett, N.; Mitchev, N.; Osman, F.; Naidoo, J.; Dorward, J.; Singh, R.; Ngobese, H.; Rompalo, A.; Mlisana, K.; Mindel, A. Diagnostic accuracy of the Xpert CT/NG and OSOM Trichomonas Rapid assays for point-of-care STI testing among young women in South Africa: A cross-sectional study. BMJ Open 2019, 9, e026888. [Google Scholar] [CrossRef] [Green Version]
- Dize, L.; Silver, B.; Gaydos, C. Comparison of the Cepheid GeneXpert CT/NG assay to the Hologic Aptima Combo2 assay for the detection of Chlamydia trachomatis and Neisseria gonorrhoeae in self-collected rectal swabs. Diagn. Microbiol. Infect. Dis. 2018, 90, 83–84. [Google Scholar] [CrossRef]
- Geiger, R.; Smith, D.M.; Little, S.J.; Mehta, S.R. Validation of the GeneXpert(R) CT/NG Assay for use with Male Pharyngeal and Rectal Swabs. Austin. J. HIV. AIDS Res. 2016, 3, 1021. [Google Scholar]
- Murtagh, M.M. The Point-of-Care Diagnostic Landscape for Sexually Transmitted Infections (STIs). WHO, 2019. Available online: https://www.who.int/reproductivehealth/topics/rtis/Diagnostic-Landscape-for-STIs-2019.pdf (accessed on 30 January 2020).
- Horst, A.L.; Rosenbohm, J.M.; Kolluri, N.; Hardick, J.; Gaydos, C.A.; Cabodi, M.; Klapperich, C.M.; Linnes, J.C. A paperfluidic platform to detect Neisseria gonorrhoeae in clinical samples. Biomed. Microdevices 2018, 20, 35. [Google Scholar] [CrossRef] [Green Version]
- Harding-Esch, E.M.; Fuller, S.S.; Chow, S.L.; Nori, A.V.; Harrison, M.A.; Parker, M.; Piepenburg, O.; Forrest, M.S.; Brooks, D.G.; Patel, R.; et al. Diagnostic accuracy of a prototype rapid chlamydia and gonorrhoea recombinase polymerase amplification assay: A multicentre cross-sectional preclinical evaluation. Clin. Microbiol. Infect. 2019, 25, 380.e1–380.e7. [Google Scholar] [CrossRef] [Green Version]
- Chin, C.D.; Laksanasopin, T.; Cheung, Y.K.; Steinmiller, D.; Linder, V.; Parsa, H.; Wang, J.; Moore, H.; Rouse, R.; Umviligihozo, G.; et al. Microfluidics-based diagnostics of infectious diseases in the developing world. Nat. Med. 2011, 17, 1015–1019. [Google Scholar] [CrossRef]
- Tsaloglou, M.N.; Nemiroski, A.; Camci-Unal, G.; Christodouleas, D.C.; Murray, L.P.; Connelly, J.T.; Whitesides, G.M. Handheld isothermal amplification and electrochemical detection of DNA in resource-limited settings. Anal. Biochem. 2018, 543, 116–121. [Google Scholar] [CrossRef]
- Unemo, M.; Shafer, W.M. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: Past, evolution, and future. Clin. Microbiol. Rev. 2014, 27, 587–613. [Google Scholar] [CrossRef] [Green Version]
- Donà, V.; Low, N.; Golparian, D.; Unemo, M. Recent advances in the development and use of molecular tests to predict antimicrobial resistance in Neisseria gonorrhoeae. Expert. Rev. Mol. Diagn. 2017, 17, 845–859. [Google Scholar] [CrossRef]
- Peterson, S.W.; Martin, I.; Demczuk, W.; Bharat, A.; Hoang, L.; Wylie, J.; Allen, V.; Lefebvre, B.; Tyrrell, G.; Horsman, G.; et al. Molecular assay for detection of ciprofloxacin resistance in Neisseria gonorrhoeae isolates from cultures and clinical nucleic acid amplification test specimens. J. Clin. Microbiol. 2015, 53, 3606–3608. [Google Scholar] [CrossRef] [Green Version]
- Pond, M.J.; Hall, C.L.; Miari, V.F.; Cole, M.; Laing, K.G.; Jagatia, H.; Harding-Esch, E.; Monahan, I.M.; Planche, T.; Hinds, J.; et al. Accurate detection of Neisseria gonorrhoeae ciprofloxacin susceptibility directly from genital and extragenital clinical samples: Towards genotype-guided antimicrobial therapy. J. Antimicrob. Chemother. 2016, 71, 897–902. [Google Scholar] [CrossRef] [Green Version]
- Siedner, M.J.; Pandori, M.; Castro, L.; Barry, P.; Whittington, W.L.; Liska, S.; Klausner, J.D. Real-time PCR assay for detection of quinolone- resistant Neisseria gonorrhoeae in urine samples. J. Clin. Microbiol. 2007, 45, 1250–1254. [Google Scholar] [CrossRef] [Green Version]
- Trembizki, E.; Buckley, C.; Donovan, B.; Chen, M.; Guy, R.; Kaldor, J.; Lahra, M.M.; Regan, D.G.; Smith, H.; Ward, J.; et al. Direct real-time PCR-based detection of Neisseria gonorrhoeae 23S rRNA mutations associated with azithromycin resistance. J. Antimicrob. Chemother. 2015, 70, 3244–3249. [Google Scholar] [CrossRef] [Green Version]
- Ochiai, S.; Ishiko, H.; Yasuda, M.; Deguchi, T. Rapid detection of the mosaic structure of the Neisseria gonorrhoeae penA gene, which is associated with decreased susceptibilities to oral cephalosporins. J. Clin. Microbiol. 2008, 46, 1804–1810. [Google Scholar] [CrossRef] [Green Version]
- Goire, N.; Freeman, K.; Lambert, S.B.; Nimmo, G.R.; Limnios, A.E.; Lahra, M.M.; Nissen, M.D.; Sloots, T.P.; Whiley, D.M. The influence of target population on nonculture-based detection of markers of Neisseria gonorrhoeae antimicrobial resistance. Sex Health 2012, 9, 422–429. [Google Scholar] [CrossRef]
- Peterson, S.W.; Martin, I.; Demczuk, W.; Bharat, A.; Hoang, L.; Wylie, J.; Lefebvre, B.; Tyrrell, G.; Horsman, G.; Haldane, D.; et al. Molecular assay for detection of genetic markers associated with decreased susceptibility to cephalosporins in Neisseria gonorrhoeae. J. Clin. Microbiol. 2015, 53, 2042–2048. [Google Scholar] [CrossRef] [Green Version]
- Trembizki, E.; Wand, H.; Donovan, B.; Chen, M.; Fairley, C.K.; Freeman, K.; Guy, R.; Kaldor, J.M.; Lahra, M.M.; Lawrence, A.; et al. The molecular epidemiology and antimicrobial resistance of Neisseria gonorrhoeae in Australia: A nationwide cross-sectional study, 2012. Clin. Infect. Dis. 2016, 63, 1591–1598. [Google Scholar] [CrossRef] [Green Version]
- Dona, V.; Kasraian, S.; Lupo, A.; Guilarte, Y.N.; Hauser, C.; Furrer, H.; Unemo, M.; Low, N.; Endimiani, A. Multiplex real-time PCR assay with high-resolution melting analysis for characterization of antimicrobial resistance in Neisseria gonorrhoeae. J. Clin. Microbiol. 2016, 54, 2074–2081. [Google Scholar] [CrossRef] [Green Version]
- Balashov, S.; Mordechai, E.; Adelson, M.E.; Gygax, S.E. Multiplex bead suspension array for screening Neisseria gonorrhoeae antibiotic resistance genetic determinants in noncultured clinical samples. J. Mol. Diagn. 2013, 15, 116–129. [Google Scholar] [CrossRef]
- Donà, V.; Smid, J.H.; Kasraian, S.; Egli-Gany, D.; Dost, F.; Imeri, F.; Unemo, M.; Low, N.; Endimiani, A. Mismatch Amplification Mutation Assay-Based Real-Time PCR for Rapid Detection of Neisseria gonorrhoeae and Antimicrobial Resistance Determinants in Clinical Specimens. J. Clin. Microbiol. 2018, 56, e00365–18. [Google Scholar] [CrossRef] [Green Version]
- Low, N.; Unemo, M. Molecular tests for the detection of antimicrobial resistant Neisseria gonorrhoeae: When, where, and how to use? Curr. Opin. Infect. Dis. 2016, 29, 45–51. [Google Scholar] [CrossRef] [Green Version]
- Goire, N.; Lahra, M.M.; Chen, M.; Donovan, B.; Fairley, C.K.; Guy, R.; Kaldor, J.; Regan, D.; Ward, J.; Nissen, M.D.; et al. Molecular approaches to enhance surveillance of gonococcal antimicrobial resistance. Nat. Rev. Microbiol. 2014, 12, 223–229. [Google Scholar] [CrossRef]
- Allan-Blitz, L.T.; Klausner, J.D. Codon 91 Gyrase A testing is necessary and sufficient to predict ciprofloxacin susceptibility in Neisseria gonorrhoeae. J. Infect. Dis. 2017, 215, 491. [Google Scholar] [CrossRef] [Green Version]
- Goire, N.; Kundu, R.; Trembizki, E.; Buckley, C.; Hogan, T.R.; Lewis, D.A.; Branley, J.M.; Whiley, D.M.; Lahra, M.M. Mixed gonococcal infections in a high-risk population, Sydney, Australia 2015: Implications for antimicrobial resistance surveillance? J. Antimicrob. Chemother. 2017, 72, 407–409. [Google Scholar] [CrossRef]
- Zhao, L.; Zhao, S. TaqMan real-time quantitative PCR assay for detection of fluoroquinolone-resistant Neisseria gonorrhoeae. Curr. Microbiol. 2012, 65, 692–695. [Google Scholar] [CrossRef]
- Golparian, D.; Donà, V.; Sánchez-Busó, L.; Foerster, S.; Harris, S.; Endimiani, A.; Low, N.; Unemo, M. Antimicrobial resistance prediction and phylogenetic analysis of Neisseria gonorrhoeae isolates using the Oxford Nanopore MinION sequencer. Sci. Rep. 2018, 8, 17596. [Google Scholar] [CrossRef] [Green Version]
- Eyre, D.W.; Golparian, D.; Unemo, M. Prediction of Minimum Inhibitory Concentrations of Antimicrobials for Neisseria gonorrhoeae Using Whole-Genome Sequencing. Methods Mol. Biol. 2019, 1997, 59–76. [Google Scholar] [CrossRef]




| Assay (Company) | Ng Targets | Cleared Specimen Types |
|---|---|---|
| Abbott RealTime CT/NG (Abbott) | Opa gene | Women: urine, swabs (vaginal, endocervical) Men: urine, urethral swab |
| cobas CT/NG (Roche) | Two different targets in the DR 9 region | Women: urine, swabs (vaginal, endocervical) Men: urine |
| APTIMA Combo 2 Assay (Hologic) | 16S-rRNA | urine swabs (vaginal, endocervical, urethral, rectal, pharyngeal) |
| BD MAX GC BD MAX CT/GC BD MAX CT/GC/TV | OpcA gene | urine (20-60mL of first morning urine recommended), swabs (vaginal endocervical) |
| BD ProbeTec Neisseria gonorrhoeae (GC) Qx Amplified DNA Assay | Pilin-gene inverting protein homologue | Women: urine, swabs (vaginal, endocervical) Men: urine, urethral swab |
| BDProbeTec ET Chlamydia trachomatis and Neisseria gonorrhoeae Amplified DNA Assays | Pilin-gene inverting protein homologue | Women: urine, endocervical swab Men: urine, urethral swab |
| Xpert CT/NG (Cepheid) | Two distinct chromosomal targets | urine swabs (vaginal, endocervical, rectal, pharyngeal) |
| binx io CT/NG Assay (binx health) | Not specified | vaginal swabs |
| Assay (Company) | Method of Amplification and Detection | Time to Result | Detected Pathogens |
|---|---|---|---|
| FTD STD 9 (FastTrack Diagnostics) | Real-time PCR Fluorescence | 3–4 h | Ct, Ng, Tv, Mg, Uu, Up, Gv, HSV1, HSV2 |
| Anyplex II STI-7 (Seegene) | Real-time PCR fluorescence and melting curve | 4–5 h | Ct, Ng, Tv, Mg, Uu, Up, Mh |
| Amplisense (Interlab Service) | Real-time PCR Fluorescence | 3–4 h | Ct, Ng, Tv, Mg |
| FilmArray STI (BioMerieux) | Nested PCR Fluorescence | 1 h | Ct, Ng, Tp, Tv, Mg, HSV1, HSV2, Uu, Hd |
| Easy Screen (Genetic Signatures) | 3-base real-time PCR (Bisufit-PCR) melting curve | 3 h | Ct, LGV, Ng, Mg, Tv, Uu, Up, Mh, GBS, Candida, Gv, HSV 1, HSV 2, VZV, Tp |
| STI Multiplex Aray (Randox Laboratories) | Real-time PCR Fluorescence | 30 min | Ct, Ng, Mg, Tv, Uu, Mh, Hd, Tp, HSV 1, HSV 2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meyer, T.; Buder, S. The Laboratory Diagnosis of Neisseria gonorrhoeae: Current Testing and Future Demands. Pathogens 2020, 9, 91. https://doi.org/10.3390/pathogens9020091
Meyer T, Buder S. The Laboratory Diagnosis of Neisseria gonorrhoeae: Current Testing and Future Demands. Pathogens. 2020; 9(2):91. https://doi.org/10.3390/pathogens9020091
Chicago/Turabian StyleMeyer, Thomas, and Susanne Buder. 2020. "The Laboratory Diagnosis of Neisseria gonorrhoeae: Current Testing and Future Demands" Pathogens 9, no. 2: 91. https://doi.org/10.3390/pathogens9020091
APA StyleMeyer, T., & Buder, S. (2020). The Laboratory Diagnosis of Neisseria gonorrhoeae: Current Testing and Future Demands. Pathogens, 9(2), 91. https://doi.org/10.3390/pathogens9020091