A Peptide Found in Human Serum, Derived from the C-Terminus of Albumin, Shows Antifungal Activity In Vitro and In Vivo
Abstract
:1. Introduction
2. Materials and Methods
2.1. Selection and Synthesis of Peptides
2.2. Evaluation of the In Vitro Candidacidal Activity of Peptides
2.3. Evaluation of the Hemolytic, Cytotoxic, and Genotoxic Activity of Selected Peptides
2.4. Circular Dichroism (CD) Spectroscopy
2.5. Evaluation of Apoptosis Induction in C. albicans Cells after Treatment with K13L
2.6. Evaluation of In Vivo Toxicity and Therapeutic Activity of Peptide
2.7. Transmission and Scanning Electron Microscopy Studies
2.8. Confocal Microscopy Studies
3. Results
3.1. In Vitro Fungicidal Activity of Selected Peptides
3.2. Hemolytic, Cytotoxic, and Genotoxic Effects of Selected Peptides
3.3. K13L Conformational State
3.4. Induction of Apoptosis in C. albicans Cells by Treatment with K13L
3.5. In Vivo Toxicity and Therapeutic Activity of K13L
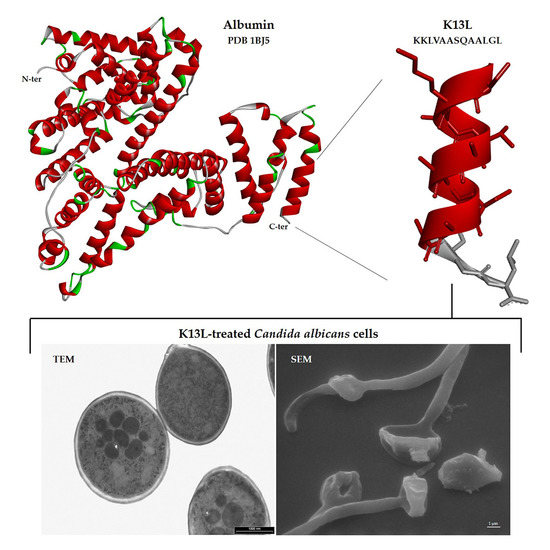
3.6. Visualization of the Effects of K13L Treatment on C. albicans Cells by Transmission and Scanning Electron Microscopy
3.7. Peptide–Yeast Cells Interaction
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization (WHO). Global Action Plan on Antimicrobial Resistance; WHO: Geneva, Switzerland, 2015. [Google Scholar]
- Zhang, Y.; Fonslow, B.R.; Shan, B.; Baek, M.C.; Yates, J.R. Protein analysis by shotgun/bottom-up proteomics. Chem. Rev. 2013, 113, 2343–2394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geho, D.H.; Liotta, L.A.; Petricoin, E.F.; Zhao, W.; Araujo, R.P. The amplified peptidome: The new treasure chest of candidate biomarkers. Curr. Opin. Chem. Biol. 2006, 10, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Khalilpour, A.; Kilic, T.; Khalilpour, S.; Álvarez, M.M.; Yazdi, I.K. Proteomic-based biomarker discovery for development of next generation diagnostics. Appl. Microbiol. Biotechnol. 2017, 101, 475–491. [Google Scholar] [CrossRef] [PubMed]
- Autelitano, D.J.; Rajic, A.; Smith, A.I.; Berndt, M.C.; Ilag, L.L.; Vadas, M. The cryptome: A subset of the proteome, comprising cryptic peptides with distinct bioactivities. Drug Discov. Today 2006, 11, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Pimenta, D.C.; Lebrun, I. Cryptides: Buried secrets in proteins. Peptides 2007, 28, 2403–2410. [Google Scholar] [CrossRef] [PubMed]
- Ueki, N.; Someya, K.; Matsuo, Y.; Wakamatsu, K.; Mukai, H. Cryptides: Functional cryptic peptides hidden in protein structures. Biopolymers 2007, 88, 190–198. [Google Scholar] [CrossRef]
- Samir, P.; Link, A.J. Analyzing the Cryptome: Uncovering Secret Sequences. AAPS J. 2011, 13, 152–158. [Google Scholar] [CrossRef]
- Iavarone, F.; Desiderio, C.; Vitali, A.; Messana, I.; Martelli, C.; Castagnola, M.; Cabras, T. Cryptides: Latent peptides everywhere. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 246–263. [Google Scholar] [CrossRef]
- Coates, C.J.; Decker, H. Immunological properties of oxygen-transport proteins: Hemoglobin, hemocyanin and hemerythrin. Cell. Mol. Life Sci. 2017, 74, 293–317. [Google Scholar] [CrossRef] [Green Version]
- Polonelli, L.; Pontón, J.; Elguezabal, N.; Moragues, M.D.; Casoli, C.; Pilotti, E.; Ronzi, P.; Dobroff, A.S.; Rodrigues, E.G.; Juliano, M.A.; et al. Antibody complementarity-determining regions (CDRs) can display differential antimicrobial, antiviral and antitumor activities. PLoS ONE 2008, 3, e2371. [Google Scholar] [CrossRef] [Green Version]
- Gabrielli, E.; Pericolini, E.; Cenci, E.; Ortelli, F.; Magliani, W.; Ciociola, T.; Bistoni, F.; Conti, S.; Vecchiarelli, A.; Polonelli, L. Antibody complementarity-determining regions (CDRs): A bridge between adaptive and innate immunity. PLoS ONE 2009, 4, e8187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polonelli, L.; Ciociola, T.; Magliani, W.; Zanello, P.P.; D’Adda, T.; Galati, S.; De Bernardis, F.; Arancia, S.; Gabrielli, E.; Pericolini, E.; et al. Peptides of the constant region of antibodies display fungicidal activity. PLoS ONE 2012, 7, e34105. [Google Scholar] [CrossRef] [Green Version]
- Gabrielli, E.; Pericolini, E.; Cenci, E.; Monari, C.; Magliani, W.; Ciociola, T.; Conti, S.; Gatti, R.; Bistoni, F.; Polonelli, L.; et al. Antibody constant region peptides can display immunomodulatory activity through activation of the Dectin-1 signalling pathway. PLoS ONE 2012, 7, e43972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polonelli, L.; Ciociola, T.; Elviri, L.; Zanello, P.P.; Giovati, L.; Arruda, D.C.; Muñoz, J.E.; Mortara, R.A.; Morace, G.; Borghi, E.; et al. A Naturally Occurring Antibody Fragment Neutralizes Infectivity of Diverse Infectious Agents. Sci. Rep. 2016, 6, 35018. [Google Scholar] [CrossRef]
- Fuchs, B.B.; O’Brien, E.; El Khoury, J.B.; Mylonakis, E. Methods for using Galleria mellonella as a model host to study fungal pathogenesis. Virulence 2010, 1, 475–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.D.; Deng, L.; Hu, G.H.; Zhao, L.X.; Hu, D.D.; Jiang, Y.Y.; Wang, Y. Using Galleria mellonella-Candida albicans infection model to evaluate antifungal agents. Biol. Pharm. Bull. 2013, 36, 1482–1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, J.; Coates, C.J.; Seoane, P.I.; Garelnabi, M.; Taylor-Smith, L.M.; Monteith, P.; Macleod, C.L.; Escaron, C.J.; Brown, G.D.; Hall, R.A.; et al. Characterizing the Mechanisms of Nonopsonic Uptake of Cryptococci by Macrophages. J. Immunol. 2018, 200, 3539–3546. [Google Scholar] [CrossRef] [PubMed]
- Emery, H.; Johnston, R.; Rowley, A.F.; Coates, C.J. Indomethacin-induced gut damage in a surrogate insect model, Galleria mellonella. Arch. Toxicol. 2019, 93, 2347–2360. [Google Scholar] [CrossRef] [Green Version]
- Coates, C.J.; Lim, J.; Harman, K.; Rowley, A.F.; Griffiths, D.J.; Emery, H.; Layton, W. The insect, Galleria mellonella, is a compatible model for evaluating the toxicology of okadaic acid. Cell. Biol. Toxicol. 2019, 35, 219–232. [Google Scholar] [CrossRef] [Green Version]
- Koomen, J.M.; Li, D.H.; Xiao, L.C.; Liu, T.C.; Coombes, K.R.; Abbruzzese, J.; Kobayashi, R. Direct tandem mass spectrometry reveals limitations in protein profiling experiments for plasma biomarker discovery. J. Proteome Res. 2005, 4, 972–981. [Google Scholar] [CrossRef]
- Williams, D.; Ackloo, S.; Zhu, P.H.; Bowden, P.; Evans, K.R.; Addison, C.L.; Lock, C.; Marshall, J.G. Precipitation and selective extraction of human serum endogenous peptides with analysis by quadrupole time-of-flight mass spectrometry reveals posttranslational modifications and low-abundance peptides. Anal. Bioanal. Chem. 2010, 396, 1223–1247. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Baker, H.; Hancock, W.S. Analysis of the low molecular weight serum peptidome using ultrafiltration and a hybrid ion trap-Fourier transform mass spectrometer. J. Chromatogr. A 2006, 1120, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Ziganshin, R.; Arapidi, G.; Azarkin, I.; Zaryadieva, E.; Alexeev, D.; Govorun, V.; Ivanov, V. New method for peptide desorption from abundant blood proteins for plasma/serum peptidome analyses by mass spectrometry. J. Proteom. 2011, 74, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Polonelli, L.; Ciociola, T.; Sperindè, M.; Giovati, L.; D’Adda, T.; Galati, S.; Travassos, L.R.; Magliani, W.; Conti, S. Fungicidal activity of peptides encoded by immunoglobulin genes. Sci. Rep. 2017, 7, 10896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Sun, J.; Xia, S.; Tian, X.; Cheserek, M.J.; Le, G. Mechanism of antifungal activity of antimicrobial peptide APP, a cell-penetrating peptide derivative, against Candida albicans: Intracellular DNA binding and cell cycle arrest. Appl. Microbiol. Biotechnol. 2016, 100, 3245–3253. [Google Scholar] [CrossRef]
- Ng, J.H.; Ilag, L.L. Cryptic protein fragments as an emerging source of peptide drugs. IDrugs 2006, 9, 343–346. [Google Scholar]
- Hancock, R.E.; Sahl, H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef]
- McGregor, D.P. Discovering and improving novel peptide therapeutics. Curr. Opin. Pharmacol. 2008, 8, 616–619. [Google Scholar] [CrossRef]
- Sato, A.K.; Viswanathan, M.; Kent, R.B.; Wood, C.R. Therapeutic peptides: Technological advances driving peptides into development. Curr. Opin. Biotechnol. 2006, 17, 638–642. [Google Scholar] [CrossRef]
- Ghosh, C.; Sarkar, P.; Issa, R.; Haldar, J. Alternatives to Conventional Antibiotics in the Era of Antimicrobial Resistance. Trends Microbiol. 2019, 27, 323–338. [Google Scholar] [CrossRef]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Peptide | Aminoacidic Sequence | Parent Protein | GRAVY Value 1 | pI | M.M. (Da) 2 | Charge 3 |
|---|---|---|---|---|---|---|
| K13L | KKLVAASQAALGL | Serum albumin | 0.792 | 10.00 | 1269.53 | 2+ |
| G17K | GLEEELQFSLGSKINVK | Complement C4-A | −0.282 | 4.79 | 1891.13 | 1− |
| S17K | SEETKENEGFTVTAEGK | Complement C3 | −1.394 | 4.32 | 1855.81 | 3− |
| D15R | DSGEGDFLAEGGGVR | Fibrinogen α chain | −0.580 | 3.92 | 1465.48 | 3− |
| D15T | DEAGSEADHEGTHST | Fibrinogen α chain | −1.607 | 4.17 | 1542.43 | 3− |
| Fungal Strain | EC50 1 (95% Confidence Intervals) mol/liter × 10−6 | |
|---|---|---|
| K13L | G17K | |
| Candida albicans SC5314 | 4.29 (3.21–5.76) | 18.46 (10.08–33.78) |
| C. albicans CA-6 | 6.28 (5.09–7.73) | 33.74 (22.80–49.93) |
| C. albicans SA40 | 3.08 (2.50–3.82) | 24.78 (19.64–31.26) |
| C. albicans AIDS68 | 2.48 (2.01–3.05) | 28.17 (25.15–31.56) |
| C. albicans UM4 | 7.43 (4.39–12.56) | 23.09 (9.28–57.42) |
| C. glabrata OMNI32 | 4.45 (4.15–4.76) | 13.95 (19.93–15.04) |
| Peptide | Hemolysis (%) | |||||
|---|---|---|---|---|---|---|
| 30 min | 120 min | |||||
| 25 µM | 50 µM | 100 µM | 25 µM | 50 µM | 100 µM | |
| K13L | 0.29 | 0.32 | 0.61 | 0.00 | 0.00 | 1.09 * |
| G17K | 0.00 | 0.03 | 0.13 | 0.00 | 0.00 | 0.00 |
| S17K | 0.00 | 0.35 | 0.80 | 0.00 | 0.26 | 1.12 * |
| D15R | 0.27 | 0.51 | 0.85 | 0.00 | 0.00 | 0.38 |
| D15T | 0.00 | 0.00 | 0.67 | 0.00 | 0.00 | 0.24 |
| Peptide | Cell Viability (%) | ||
|---|---|---|---|
| 25 µM | 50 µM | 100 µM | |
| K13L | 105.95 ± 1.47 | 105.04 ± 0.20 | 101.26 ± 0.38 |
| G17K | 99.07 ± 1.57 | 99.73 ± 0.45 | 99.07 ± 1.57 |
| S17K | 103.47 ± 2.00 | 102.69 ± 1.37 | 101.73 ± 1.82 |
| D15R | 101.32 ± 1.92 | 101.25 ± 0.19 | 103.00 ± 1.49 |
| D15T | 108.50 ± 0.92 | 107.55 ± 1.48 | 108.50 ± 5.15 |
| Peptide | Tail Intensity (%) | |||
|---|---|---|---|---|
| 0 | 5 µM | 10 µM | 20 µM | |
| K13L | 0.37 ± 0.04 | 0.69 ± 0.03 * | 0.59 ± 0.09 * | 0.36 ± 0.22 |
| G17K | 0.37 ± 0.04 | 0.40 ± 0.02 | 0.34 ± 0.25 | 0.12 ± 0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciociola, T.; Zanello, P.P.; D’Adda, T.; Galati, S.; Conti, S.; Magliani, W.; Giovati, L. A Peptide Found in Human Serum, Derived from the C-Terminus of Albumin, Shows Antifungal Activity In Vitro and In Vivo. Microorganisms 2020, 8, 1627. https://doi.org/10.3390/microorganisms8101627
Ciociola T, Zanello PP, D’Adda T, Galati S, Conti S, Magliani W, Giovati L. A Peptide Found in Human Serum, Derived from the C-Terminus of Albumin, Shows Antifungal Activity In Vitro and In Vivo. Microorganisms. 2020; 8(10):1627. https://doi.org/10.3390/microorganisms8101627
Chicago/Turabian StyleCiociola, Tecla, Pier Paolo Zanello, Tiziana D’Adda, Serena Galati, Stefania Conti, Walter Magliani, and Laura Giovati. 2020. "A Peptide Found in Human Serum, Derived from the C-Terminus of Albumin, Shows Antifungal Activity In Vitro and In Vivo" Microorganisms 8, no. 10: 1627. https://doi.org/10.3390/microorganisms8101627
APA StyleCiociola, T., Zanello, P. P., D’Adda, T., Galati, S., Conti, S., Magliani, W., & Giovati, L. (2020). A Peptide Found in Human Serum, Derived from the C-Terminus of Albumin, Shows Antifungal Activity In Vitro and In Vivo. Microorganisms, 8(10), 1627. https://doi.org/10.3390/microorganisms8101627