Rapid Identification of Escherichia coli Colistin-Resistant Strains by MALDI-TOF Mass Spectrometry
Abstract
:1. Introduction
2. Materials and Methods
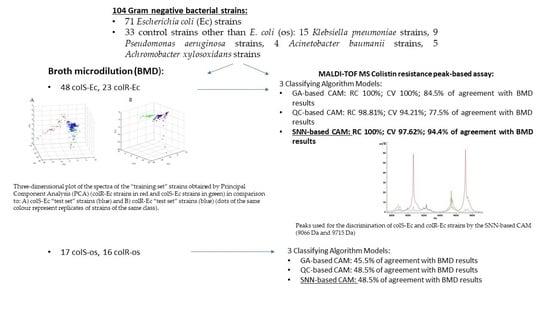
2.1. Bacterial Strains
2.2. Colistin Susceptibility Testing
2.3. MALDI-TOF MS Protein Peak-Based Assay
2.4. Classifying Algorithm Model
2.4.1. Genetic Algorithm
2.4.2. Quick Classifier
2.4.3. Supervised Neural Network
2.5. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lim, L.M.; Ly, N.; Anderson, D.; Yang, J.C.; Macander, L.; Jarkowski, A., 3rd; Forrest, A.; Bulitta, J.B.; Tsuji, B.T. Resurgence of colistin: A review of resistance, toxicity, pharmacodynamics, and dosing. Pharmacotherapy 2010, 30, 1279–1291. [Google Scholar] [CrossRef] [PubMed]
- WHO. Global Report on Surveillance 2014; 2014 AMR Rep; WHO: Geneva, Switzerland, 2014; pp. 1–8. [Google Scholar]
- El-Sayed Ahmed, M.A.E.-G.; Zhong, L.-L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.-B. Colistin and its role in the Era of antibiotic resistance: An extended review (2000–2019). Emerg. Microbes Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levy, S.B.; Marshall, B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. 2004, 10, S122–S129. [Google Scholar] [CrossRef] [PubMed]
- Christaki, E.; Marcou, M.; Tofarides, A. Antimicrobial Resistance in Bacteria: Mechanisms, Evolution, and Persistence. J. Mol. Evol. 2020, 88, 26–40. [Google Scholar] [CrossRef]
- Sherry, N.; Howden, B. Emerging Gram negative resistance to last-line antimicrobial agents fosfomycin, colistin and ceftazidime-avibactam—Epidemiology, laboratory detection and treatment implications. Expert Rev. Anti. Infect. Ther. 2018, 16, 289–306. [Google Scholar] [CrossRef] [PubMed]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falagas, M.E.; Kasiakou, S.K. Colistin: The revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2005, 40, 1333–1341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, J.M.; Dorman, D.C.; Roy, L.P. Acute renal failure due to overdosage of colistin. Med. J. Aust. 1970, 2, 923–924. [Google Scholar] [CrossRef] [PubMed]
- Koch-Weser, J.; Sidel, V.W.; Federman, E.B.; Kanarek, P.; Finer, D.C.; Eaton, A.E. Adverse effects of sodium colistimethate. Manifestations and specific reaction rates during 317 courses of therapy. Ann. Intern. Med. 1970, 72, 857–868. [Google Scholar] [CrossRef]
- Ryan, K.J.; Schainuck, L.I.; Hickman, R.O.; Striker, G.E. Colistimethate toxicity. Report of a fatal case in a previously healthy child. JAMA 1969, 207, 2099–2101. [Google Scholar] [CrossRef] [PubMed]
- Bialvaei, A.Z.; Samadi Kafil, H. Colistin, mechanisms and prevalence of resistance. Curr. Med. Res. Opin. 2015, 31, 707–721. [Google Scholar] [CrossRef]
- Hamel, M.; Rolain, J.-M.; Baron, S.A. The History of Colistin Resistance Mechanisms in Bacteria: Progress and Challenges. Microorganisms 2021, 9, 442. [Google Scholar] [CrossRef] [PubMed]
- Olaitan, A.O.; Morand, S.; Rolain, J.-M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Roh, K.H.; Hong, S.G.; Shin, H.B.; Jeong, S.H.; Song, W.; Uh, Y.; Yong, D.; Lee, K. In Vitro Synergistic Effects of Antimicrobial Combinations on Extensively Drug-Resistant Pseudomonas aeruginosa and Acinetobacter baumannii Isolates. Ann. Lab. Med. 2016, 36, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Ling, Z.; Yin, W.; Shen, Z.; Wang, Y.; Shen, J.; Walsh, T.R. Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J. Antimicrob. Chemother. 2020, 75, 3087–3095. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Wang, C.; Feng, Y.; Liu, L.; Wei, L.; Kang, M.; Zong, Z. Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 2020, 9, 508–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; van Dorp, L.; Shaw, L.P.; Bradley, P.; Wang, Q.; Wang, X.; Jin, L.; Zhang, Q.; Liu, Y.; Rieux, A.; et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 2018, 9, 1179. [Google Scholar] [CrossRef] [Green Version]
- Dolejska, M.; Papagiannitsis, C.C. Plasmid-mediated resistance is going wild. Plasmid 2018, 99, 99–111. [Google Scholar] [CrossRef]
- Jayol, A.; Nordmann, P.; Lehours, P.; Poirel, L.; Dubois, V. Comparison of methods for detection of plasmid-mediated and chromosomally encoded colistin resistance in Enterobacteriaceae. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2018, 24, 175–179. [Google Scholar] [CrossRef] [Green Version]
- Jayol, A.; Nordmann, P.; Brink, A.; Villegas, M.-V.; Dubois, V.; Poirel, L. High-Level Resistance to Colistin Mediated by Various Mutations in the crrB Gene among Carbapenemase-Producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donà, V.; Bernasconi, O.J.; Kasraian, S.; Tinguely, R.; Endimiani, A. A SYBR® Green-based real-time PCR method for improved detection of mcr-1-mediated colistin resistance in human stool samples. J. Glob. Antimicrob. Resist. 2017, 9, 57–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osei Sekyere, J. Mcr colistin resistance gene: A systematic review of current diagnostics and detection methods. Microbiologyopen 2019, 8, e00682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefaniuk, E.M.; Tyski, S. Colistin Resistance in Enterobacterales Strains—A Current View. Polish J. Microbiol. 2019, 68, 417–427. [Google Scholar] [CrossRef] [Green Version]
- Osei Sekyere, J.; Govinden, U.; Bester, L.A.; Essack, S.Y. Colistin and tigecycline resistance in carbapenemase-producing Gram-negative bacteria: Emerging resistance mechanisms and detection methods. J. Appl. Microbiol. 2016, 121, 601–617. [Google Scholar] [CrossRef] [PubMed]
- Chew, K.L.; La, M.-V.; Lin, R.T.P.; Teo, J.W.P. Colistin and Polymyxin B Susceptibility Testing for Carbapenem-Resistant and mcr-Positive Enterobacteriaceae: Comparison of Sensititre, MicroScan, Vitek 2, and Etest with Broth Microdilution. J. Clin. Microbiol. 2017, 55, 2609–2616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flores-Treviño, S.; Garza-González, E.; Mendoza-Olazarán, S.; Morfín-Otero, R.; Camacho-Ortiz, A.; Rodríguez-Noriega, E.; Martínez-Meléndez, A.; Bocanegra-Ibarias, P. Screening of biomarkers of drug resistance or virulence in ESCAPE pathogens by MALDI-TOF mass spectrometry. Sci. Rep. 2019, 9, 18945. [Google Scholar] [CrossRef] [Green Version]
- Charretier, Y.; Schrenzel, J. Mass spectrometry methods for predicting antibiotic resistance. Proteomics. Clin. Appl. 2016, 10, 964–981. [Google Scholar] [CrossRef]
- Calderaro, A.; Buttrini, M.; Piergianni, M.; Montecchini, S.; Martinelli, M.; Covan, S.; Piccolo, G.; Medici, M.C.; Arcangeletti, C.; Chezzi, C.; et al. Evaluation of a modified meropenem hydrolysis assay on a large cohort of KPC and VIM carbapenemase-producing Enterobacteriaceae. PLoS ONE 2017, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Furniss, R.C.D.; Dortet, L.; Bolland, W.; Drews, O.; Sparbier, K.; Bonnin, R.A.; Filloux, A.; Kostrzewa, M.; Mavridou, D.A.I.; Larrouy-Maumus, G. Detection of Colistin Resistance in Escherichia coli by Use of the MALDI Biotyper Sirius Mass Spectrometry System. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dortet, L.; Potron, A.; Bonnin, R.A.; Plesiat, P.; Naas, T.; Filloux, A.; Larrouy-Maumus, G. Rapid detection of colistin resistance in Acinetobacter baumannii using MALDI-TOF-based lipidomics on intact bacteria. Sci. Rep. 2018, 8, 16910. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; Lin, J.; Jia, H.; Zhou, C.; Zhang, Y.; Lin, Y.; Ye, J.; Cao, J.; Zhou, T. Resistance and Heteroresistance to Colistin in Escherichia coli Isolates from Wenzhou, China. Infect. Drug Resist. 2020, 13, 3551–3561. [Google Scholar] [CrossRef] [PubMed]
- Croxatto, A.; Prod’hom, G.; Greub, G. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol. Rev. 2012, 36, 380–407. [Google Scholar] [CrossRef]
- Calderaro, A.; Buttrini, M.; Martinelli, M.; Farina, B.; Moro, T.; Montecchini, S.; Arcangeletti, M.C.; Chezzi, C.; De Conto, F. Rapid classification of clostridioides difficile strains using maldi-tof ms peak-based assay in comparison with pcr-ribotyping. Microorganisms 2021, 9, 661. [Google Scholar] [CrossRef]
- Calderaro, A.; Motta, F.; Montecchini, S.; Gorrini, C.; Piccolo, G.; Piergianni, M.; Buttrini, M.; Medici, M.C.; Arcangeletti, M.C.; Chezzi, C.; et al. Identification of Dermatophyte Species after implementation of the in-house MALDI-TOF MS database. Int. J. Mol. Sci. 2014, 15, 16012–16024. [Google Scholar] [CrossRef] [PubMed]
- Calderaro, A.; Gorrini, C.; Piccolo, G.; Montecchini, S.; Buttrini, M.; Rossi, S.; Piergianni, M.; Arcangeletti, M.C.; De Conto, F.; Chezzi, C.; et al. Identification of Borrelia species after creation of an in-house MALDI-TOF MS database. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]



| Algorithms Model | RC (%) | CV (%) | Peaks Used for Classification (Da) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4177 | 4365 | 4440 | 4449 | 4498 | 5612 | 6257 | 6283 | 8330 | 8878 | 9066 | 9715 | |||
| GA | 100 | 100 | + | + | + | + | + | + | + | |||||
| SNN | 100 | 97.62 | + | + | + | + | + | + | + | |||||
| QC | 98.81 | 94.21 | + | + | + | |||||||||
| Set of Strains | BMD | No. | CAM Classification | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GA | SNN | QC | |||||||||
| colR | colS | Correctly Classified (%) | colR | colS | Correctly Classified (%) | colR | colS | Correctly Classified (%) | |||
| Internal validation | colR | 5 | 5 | 5/5 (100) | 5 | 5/5 (100) | 5 | 5/5 (100) | |||
| colS | 5 | 5 | 5/5 (100) | 5 | 5/5 (100) | 5 | 5/5 (100) | ||||
| 10 | 10/10 (100) | 10/10 (100) | 10/10 (100) | ||||||||
| External validation | colR | 18 | 16 | 2 | 16/18 (88.9) | 15 | 3 | 15/18 (83.3) | 17 | 1 | 17/18 (94.4) |
| colS | 43 | 9 | 34 | 34/43 (79) | 1 | 42 | 42/43 (97.7) | 15 | 28 | 28/43 (65.1) | |
| 61 | 50/61 (82) | 57/61 (93.4) | 45/61 (73.8) | ||||||||
| Overall | 71 | 60/71 (84.5) | 67/71 (94.4) | 55/71 (77.5) | |||||||
| Strains | BMD | No. | CAM Classification | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GA | SNN | QC | |||||||||
| colR | colS | Correctly Classified (%) | colR | colS | Correctly Classified (%) | colR | colS | Correctly Classified (%) | |||
| K. pneumoniae | colR | 7 | 3 | 4 | 3/7 (42.9) | 7 | 0/7 (0) | 7 | 7/7 (100) | ||
| P. aeruginosa | colR | 2 | 2 | 2/2 (100) | 2 | 2/2 (100) | 2 | 2/2 (100) | |||
| A. baumannii | colR | 2 | 2 | 2/2 (100) | 2 | 2/2 (100) | 2 | 2/2 (100) | |||
| A. xylosoxydans | colR | 5 | 1 | 4 | 1/5 (20) | 4 | 1 | 4/5 (80) | 5 | 5/5 (100) | |
| 16 | 8/16 (50) | 8/16 (50) | 16/16 (100) | ||||||||
| K. pneumoniae | colS | 8 | 1 | 7 | 7/8 (87.5) | 8 | 8/8 (100) | 8 | 0/8 (0) | ||
| P. aeruginosa | colS | 7 | 7 | 0/7 (0) | 7 | 0/7 (0) | 7 | 0/7 (0) | |||
| A. baumannii | colS | 2 | 2 | 0/2 (0) | 2 | 0/2(0) | 2 | 0/2 (0) | |||
| 17 | 7/17 (41.2) | 8/17 (47) | 0/17 (0) | ||||||||
| Overall | 33 | 15/33 (45.5) | 16/33 (48.5) | 16/33 (48.5) | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calderaro, A.; Buttrini, M.; Farina, B.; Montecchini, S.; Martinelli, M.; Crocamo, F.; Arcangeletti, M.C.; Chezzi, C.; De Conto, F. Rapid Identification of Escherichia coli Colistin-Resistant Strains by MALDI-TOF Mass Spectrometry. Microorganisms 2021, 9, 2210. https://doi.org/10.3390/microorganisms9112210
Calderaro A, Buttrini M, Farina B, Montecchini S, Martinelli M, Crocamo F, Arcangeletti MC, Chezzi C, De Conto F. Rapid Identification of Escherichia coli Colistin-Resistant Strains by MALDI-TOF Mass Spectrometry. Microorganisms. 2021; 9(11):2210. https://doi.org/10.3390/microorganisms9112210
Chicago/Turabian StyleCalderaro, Adriana, Mirko Buttrini, Benedetta Farina, Sara Montecchini, Monica Martinelli, Federica Crocamo, Maria Cristina Arcangeletti, Carlo Chezzi, and Flora De Conto. 2021. "Rapid Identification of Escherichia coli Colistin-Resistant Strains by MALDI-TOF Mass Spectrometry" Microorganisms 9, no. 11: 2210. https://doi.org/10.3390/microorganisms9112210
APA StyleCalderaro, A., Buttrini, M., Farina, B., Montecchini, S., Martinelli, M., Crocamo, F., Arcangeletti, M. C., Chezzi, C., & De Conto, F. (2021). Rapid Identification of Escherichia coli Colistin-Resistant Strains by MALDI-TOF Mass Spectrometry. Microorganisms, 9(11), 2210. https://doi.org/10.3390/microorganisms9112210