Biosensors Used for Epifluorescence and Confocal Laser Scanning Microscopies to Study Dickeya and Pectobacterium Virulence and Biocontrol
Abstract
:1. Introduction
2. Main Traits of Fluorescent Proteins Used to Construct Tagged-Soft-Rot Pectobacteriaceae
2.1. Fluorescent Proteins: Origin, Structure, Spectral Range, and Stability
2.2. Interests of Fluorescent Proteins for Studies of Plant-Bacteria Interactions
3. Colonization of Plants by Fluorescent Protein-Tagged Dickeya spp. and Pectobacterium spp.
4. Visualization of Biocontrol Strategies to Counteract Virulence of Dickeya and Pectobacterium in Potato Plant and Tuber
4.1. Plant Root Colonization and Biocontrol Effectiveness of Fluorescent-Tagged Soft-Rot Pectobacteriaceae Antagonists
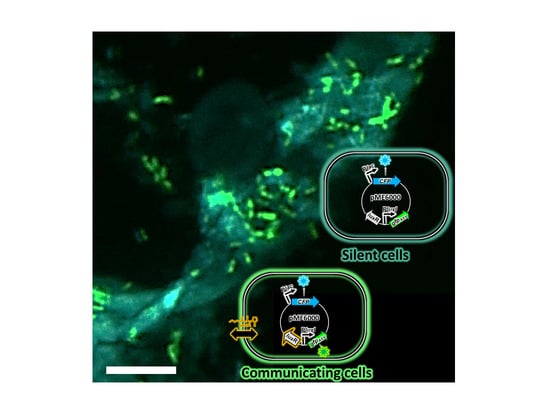
4.2. Tracking and Interference of Quorum-Sensing Communication Used by Soft-Rot Pectobacteria
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adeolu, M.; Alnajar, S.; Naushad, S.S.; Gupta, R. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: Proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5575–5599. [Google Scholar] [CrossRef] [PubMed]
- Hélias, V.; Quêtu-Laurent, A.; Gauthier, J.-P.; Le-Hingrat, Y.; Andrivon, D. Screening Dickeya pathogenicity to potato shows inter- and intraspecific diversity. Potato Res. 2014, 57, 175. [Google Scholar] [CrossRef]
- Ma, B.; Hibbing, M.E.; Kim, H.-S.; Reedy, R.M.; Yedidia, I.; Breuer, J.; Breuer, J.; Glasner, J.D.; Perna, N.T.; Kelman, A.; et al. Host range and molecular phylogenies of the soft rot enterobacterial genera Pectobacterium and Dickeya. Phytopathology 2007, 97, 1150–1163. [Google Scholar] [CrossRef] [Green Version]
- Reverchon, S.; Nasser, W. Dickeya ecology, environment sensing and regulation of virulence programme: Dickeya dadantii pathogenicity. Environ. Microbiol. Rep. 2013, 5, 622–636. [Google Scholar] [CrossRef] [PubMed]
- Oulghazi, S.; Moumni, M.; Khayi, S.; Robic, K.; Sarfraz, S.; Lopez-Roques, C.; Vandecasteele, C.; Faure, D. Dickeya dianthicola emerged from a potato culture area exhibiting a wide diversity of pectinolytic pathogens in northern Morocco. BMC Genom. 2020. preprint in review. [Google Scholar] [CrossRef]
- Oulghazi, S.; Pédron, J.; Cigna, J.; Lau, Y.Y.; Moumni, M.; Van Gijsegem, F.; Chan, K.-G.; Faure, D. Dickeya undicola sp. nov., a novel species for pectinolytic isolates from surface waters in Europe and Asia. Int. J. Syst. Evol. Microbiol. 2019, 69, 2440–2444. [Google Scholar] [CrossRef]
- Pasanen, M.; Waleron, M.; Schott, T.; Cleenwerck, I.; Misztak, A.; Waleron, K.; Pritchard, L.; Bakr, R.; Degefu, Y.; van der Wolf, J.; et al. Pectobacterium parvum sp. nov., having a Salmonella SPI-1-like Type III secretion system and low virulence. Int. J. Syst. Evol. Microbiol. 2020, 70, 2440–2448. [Google Scholar] [CrossRef]
- Portier, P.; Pédron, J.; Taghouti, G.; Fischer-Le Saux, M.; Caullireau, E.; Bertrand, C.; Laurent, A.; Chawki, K.; Oulgazi, S.; Moumni, M.; et al. Elevation of Pectobacterium carotovorum subsp. odoriferum to species level as Pectobacterium odoriferum sp. nov., proposal of Pectobacterium brasiliense sp. nov. and Pectobacterium actinidiae sp. nov., emended description of Pectobacterium carotovorum and description of Pectobacterium versatile sp. nov., isolated from streams and symptoms on diverse plants. Int. J. Syst. Evol. Microbiol. 2019, 69, 3207–3216. [Google Scholar] [CrossRef]
- Sarfraz, S.; Sahi, S.T.; Oulghazi, S.; Riaz, K.; Rajput, N.A.; Atiq, M.; Tufail, M.R.; Hameed, A.; Faure, D. Species diversity of Dickeya and Pectobacterium causing potato blackleg disease in Pakistan. Plant Dis. 2020, 104, 1492–1499. [Google Scholar] [CrossRef]
- Toth, I.K.; van der Wolf, J.M.; Saddler, G.; Lojkowska, E.; Hélias, V.; Pirhonen, M.; Tsror Lahkim, L.; Elphinstone, J.G. Dickeya species: An emerging problem for potato production in Europe: Dickeya spp. on potato in Europe. Plant Pathol. 2011, 60, 385–399. [Google Scholar] [CrossRef]
- Van der Wolf, J.; Nijhuis, E.H.; Kowaleswska, M.J.; Saddler, G.S.; Parkinson, N.; Elphinstone, J.G.; Pritchard, L.; Toth, I.K.; Lojkowska, E.; Potrykus, M.; et al. Dickeya solani sp. nov. a pectinolytic plant-pathogenic bacterium isolated from potato (Solanum tuberosum). Int. J. Syst. Evol. Microbiol. 2014, 64, 768–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waleron, M.; Misztak, A.; Waleron, M.; Franczuk, M.; Jońca, J.; Wielgomas, B.; Mikiciński, A.; Popović, T.; Waleron, K. Pectobacterium zantedeschiae sp. nov. a new species of a soft rot pathogen isolated from Calla lily (Zantedeschia spp.). Syst. Appl. Microbiol. 2019, 42, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Waleron, M.; Misztak, A.; Waleron, M.; Jonca, J.; Furmaniak, M.; Waleron, K. Pectobacterium polonicum sp. nov. isolated from vegetable fields. Int. J. Syst. Evol. Microbiol. 2019, 69, 1751–1759. [Google Scholar] [CrossRef]
- Czajkowski, R.; Pérombelon, M.C.M.; Jafra, S.; Lojkowska, E.; Potrykus, M.; van der Wolf, J.M.; Sledz, W. Detection, identification and differentiation of Pectobacterium and Dickeya species causing potato blackleg and tuber soft rot: A review. Ann. Appl. Biol. 2015, 166, 18–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hugouvieux-Cotte-Pattat, N.; Jacot-des-Combes, C.; Briolay, J. Dickeya lacustris sp. nov., a water-living pectinolytic bacterium isolated from lakes in France. Int. J. Syst. Evol. Microbiol. 2019, 69, 721–726. [Google Scholar] [CrossRef]
- Pédron, J.; Van Gijsegem, F. Diversity in the bacterial genus Dickeya grouping plant pathogens and waterways isolates. OBM Genet. 2019, 3, 22. [Google Scholar] [CrossRef] [Green Version]
- Glasner, J.D.; Marquez-Villavicencio, M.; Kim, H.-S.; Jahn, C.E.; Ma, B.; Biehl, B.S.; Rissman, A.I.; Mole, B.; Yi, X.; Yang, C.-H.; et al. Niche-specificity and the variable fraction of the Pectobacterium pan-genome. Mol. Plant Microbe Interact. 2008, 21, 1549–1560. [Google Scholar] [CrossRef]
- Grenier, A.M.; Duport, G.; Pagès, S.; Condemine, G.; Rahbé, Y. The phytopathogen Dickeya dadantii (Erwinia chrysanthemi 3937) is a pathogen of the pea aphid. Appl. Environ. Microbiol. 2006, 72, 1956–1965. [Google Scholar] [CrossRef] [Green Version]
- Rossmann, S.; Dees, M.W.; Perminow, J.; Meadow, R.; Brurberg, M.B. Soft rot enterobacteriaceae are carried by a large range of insect species in potato fields. Appl. Environ. Microbiol. 2018, 84, e00281-18. [Google Scholar] [CrossRef] [Green Version]
- Charkowski, A.O. The changing face of bacterial soft-rot diseases. Annu. Rev. Phytopathol. 2018, 56, 269–288. [Google Scholar] [CrossRef]
- FAO (Food and Agriculture Organization). International Year of the Potato 2008, New Light on a Hidden Treasure; Food and Agriculture Organization of the United Nations: Rome, Italy, 2009; p. 144. Available online: http://www.fao.org/3/i0500e/i0500e00.htm (accessed on 12 May 2020).
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hélias, V.; Andrivon, D.; Jouan, B. Development of symptoms caused by Erwinia carotovora ssp. atroseptica under field conditions and their effects on the yield of individual potato plants. Plant Pathol. 2000, 49, 23–32. [Google Scholar] [CrossRef]
- Hélias, V.; Andrivon, D.; Jouan, B. Internal colonization pathways of potato plants by Erwinia carotovora ssp. atroseptica. Plant Pathol. 2000, 49, 33–42. [Google Scholar] [CrossRef]
- Pérombelon, M.C.M. Potato diseases caused by soft rot erwinias: An overview of pathogenesis. Plant Pathol. 2002, 51, 1–12. [Google Scholar] [CrossRef]
- Czajkowski, R.; Pérombelon, M.C.M.; van Veen, J.A.; van der Wolf, J.M. Control of blackleg and tuber soft rot of potato caused by Pectobacterium and Dickeya species: A review. Plant Pathol. 2011, 60, 999–1013. [Google Scholar] [CrossRef]
- Skelsey, P.; Elphinstone, J.G.; Saddler, G.S.; Wale, S.J.; Toth, I.K. Spatial analysis of blackleg-affected seed potato crops in Scotland. Plant Pathol. 2016, 65, 570–576. [Google Scholar] [CrossRef]
- Gorshkov, V.Y.; Daminova, A.G.; Mikshina, P.V.; Petrova, O.E.; Ageeva, M.V.; Salnikov, V.V.; Gorshkova, T.A.; Gogolev, Y.V. Pathogen-induced conditioning of the primary xylem vessels—A prerequisite for the formation of bacterial emboli by Pectobacterium atrosepticum. Plant Biol. 2016, 18, 609–617. [Google Scholar] [CrossRef]
- Moleleki, L.N.; Pretorius, R.G.; Tanui, C.K.; Mosina, G.; Theron, J. A quorum sensing-defective mutant of Pectobacterium carotovorum ssp. brasiliense 1692 is attenuated in virulence and unable to occlude xylem tissue of susceptible potato plant stems. Mol. Plant Pathol. 2017, 18, 32–44. [Google Scholar] [CrossRef] [Green Version]
- Hugouvieux-Cotte-Pattat, N. Metabolism and virulence strategies in Dickeya-host interactions. Prog. Mol. Biol. Transl. Sci. 2016, 142, 93–129. [Google Scholar] [CrossRef]
- Smadja, B.; Latour, X.; Trigui, S.; Burini, J.-F.; Chevalier, S.; Orange, N. Thermodependence of growth and enzymatic activities implicated in pathogenicity of two Erwinia carotovora subspecies (Pectobacterium spp.). Can. J. Microbiol. 2004, 50, 19–27. [Google Scholar] [CrossRef]
- Bowden, S.D.; Eyres, A.; Chung, J.C.S.; Monson, R.E.; Thompson, A.; Salmond, G.P.C.; Spring, D.R.; Welch, M. Virulence in Pectobacterium atrosepticum is regulated by a coincidence circuit involving quorum sensing and the stress alarmone, (p)ppGpp: (p)ppGpp regulates rsmB expression. Mol. Microbiol. 2013, 90, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Crépin, A.; Barbey, C.; Beury-Cirou, A.; Hélias, V.; Taupin, L.; Reverchon, S.; Nasser, W.; Faure, D.; Dufour, A.; Orange, N.; et al. Quorum sensing signaling molecules produced by reference and emerging soft-rot bacteria (Dickeya and Pectobacterium spp.). PLoS ONE 2012, 7, e35176. [Google Scholar] [CrossRef] [PubMed]
- Crépin, A.; Beury-Cirou, A.; Barbey, C.; Farmer, C.; Hélias, V.; Burini, J.-F.; Faure, D.; Latour, X. N-Acyl homoserine lactones in diverse Pectobacterium and Dickeya plant pathogens: Diversity, abundance, and involvement in virulence. Sensors 2012, 12, 3484–3497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnard, A.M.L.; Salmond, G.P.C. Quorum sensing in Erwinia species. Anal. Bioanal. Chem. 2007, 387, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Latour, X.; Diallo, S.; Chevalier, S.; Morin, D.; Smadja, B.; Burini, J.F.; Haras, D.; Orange, N. Thermoregulation of N-acyl homoserine lactone-based quorum sensing in the soft rot bacterium Pectobacterium atrosepticum. Appl. Environ. Microbiol. 2007, 73, 4078–4081. [Google Scholar] [CrossRef] [Green Version]
- Põllumaa, L.; Alamäe, T.; Mäe, A. Quorum sensing and expression of virulence in Pectobacteria. Sensors 2012, 12, 3327–3349. [Google Scholar] [CrossRef] [Green Version]
- Nasser, W.; Dorel, C.; Wawrzyniak, J.; Van Gijsegem, F.; Groleau, M.-C.; Déziel, E.; Reverchon, S. Vfm a new quorum sensing system controls the virulence of Dickeya dadantii. Environ. Microbiol. 2013, 15, 865–880. [Google Scholar] [CrossRef]
- Smadja, B.; Latour, X.; Faure, D.; Chevalier, S.; Dessaux, Y.; Orange, N. Involvement of N-acylhomoserine lactones throughout plant infection by Erwinia carotovora subsp. atroseptica (Pectobacterium atrosepticum). Mol. Plant Microbe. Interact. 2004, 17, 1269–1278. [Google Scholar] [CrossRef] [Green Version]
- Chudakov, D.M.; Matz, M.V.; Lukyanov, S.; Lukyanov, K.A. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol. Rev. 2010, 90, 1103–1163. [Google Scholar] [CrossRef]
- Shimomura, O.; Johnson, F.H.; Saiga, Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J. Cell Comp. Physiol. 1962, 59, 223–239. [Google Scholar] [CrossRef]
- Tsien, R.Y. The green fluorescent protein. Annu. Rev. Biochem. 1998, 67, 509–544. [Google Scholar] [CrossRef] [PubMed]
- Chalfie, M.; Tu, Y.; Euskirchen, G.; Ward, W.W.; Prasher, D.C. Green fluorescent protein as a marker for gene expression. Science 1994, 263, 802–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, J.B.; Sternberg, C.; Poulsen, L.K.; Bjørn, S.P.; Givskov, M.; Molin, S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 1998, 64, 2240–2246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, J.B.; Heydorn, A.; Hentzer, M.; Eberl, L.; Geisenberger, O.; Christensen, B.B.; Molin, S.; Givskov, M. Gfp-based N-acyl homoserine-lactone sensor systems for detection of bacterial communication. Appl. Environ. Microbiol. 2001, 67, 575–585. [Google Scholar] [CrossRef] [Green Version]
- Steidle, A.; Sigl, K.; Schuhegger, R.; Ihring, A.; Schmid, M.; Gantner, S.; Stoffels, M.; Riedel, K.; Givskov, M.; Hartmann, A.; et al. Visualization of N-acylhomoserine lactone- mediated cell-cell communication between bacteria colonizing the tomato rhizosphere. Appl. Environ. Microbiol. 2001, 67, 5761–5770. [Google Scholar] [CrossRef] [Green Version]
- Battistutta, R.; Negro, A.; Zanotti, G. Crystal structure and refolding properties of the mutant F99S/M153T/V163A of the green fluorescent protein. Proteins 2000, 41, 429–437. [Google Scholar] [CrossRef]
- Remington, S.J. Green fluorescent protein: A perspective. Protein Sci. 2011, 20, 1509–1519. [Google Scholar] [CrossRef] [Green Version]
- Errampalli, D.; Leung, K.; Cassidy, M.B.; Kostrzynska, M.; Blears, M.; Lee, H.; Trevors, J.T. Applications of the green fluorescent protein as a molecular marker in environmental microorganisms. J. Microbiol. Methods 1999, 35, 187–199. [Google Scholar] [CrossRef]
- Heim, R.; Tsien, R.Y. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr. Biol. 1996, 6, 178–182. [Google Scholar] [CrossRef] [Green Version]
- Kremers, G.J.; Goedhart, J.; van den Heuvel, D.J.; Gerritsen, H.C.; Gadella, T.W., Jr. Improved green and blue fluorescent proteins for expression in bacteria and mammalian cells. Biochemistry 2007, 46, 3775–3783. [Google Scholar] [CrossRef]
- Kremers, G.J.; Goedhart, J.; van Munster, E.B.; Gadella, T.W., Jr. Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Förster radius. Biochemistry 2006, 45, 6570–6580. [Google Scholar] [CrossRef] [PubMed]
- Labas, Y.A.; Gurskaya, N.G.; Yanushevich, Y.G.; Fradkov, A.F.; Lukyanov, K.A.; Lukyanov, S.A.; Matz, M.V. Diversity and evolution of the green fluorescent protein family. Proc. Natl. Acad. Sci. USA 2002, 99, 4256–4261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, J.N.; Ai, H.W.; Campbell, R.E.; Remington, S.J. Structural basis for reversible photobleaching of a green fluorescent protein homologue. Proc. Natl. Acad. Sci. USA 2007, 104, 6672–6677. [Google Scholar] [CrossRef] [Green Version]
- Matz, M.V.; Fradkov, A.F.; Labas, Y.A.; Savitsky, A.P.; Zaraisky, A.G.; Markelov, M.L.; Lukyanov, S.A. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 1999, 17, 969–973. [Google Scholar] [CrossRef] [PubMed]
- Shagin, D.A.; Barsova, E.V.; Yanushevich, Y.G.; Fradkov, A.F.; Lukyanov, K.A.; Labas, Y.A.; Semenova, T.N.; Ugalde, J.A.; Meyers, A.; Nunez, J.M.; et al. GFP-like proteins as ubiquitous metazoan superfamily: Evolution of functional features and structural complexity. Mol. Biol. Evol. 2004, 21, 841–850. [Google Scholar] [CrossRef]
- Andresen, M.; Stiel, A.C.; Fölling, J.; Wenzel, D.; Schönle, A.; Egner, A.; Eggeling, C.; Hell, S.W.; Jakobs, S. Photoswitchable fluorescent proteins enable monochromatic multilabel imaging and dual color fluorescence nanoscopy. Nat. Biotechnol. 2008, 26, 1035–1040. [Google Scholar] [CrossRef]
- Adam, V.; Lelimousin, M.; Boehme, S.; Desfond, G.; Nienhaus, K.; Field, M.J.; Wiedenmann, J.; McSweeney, S.; Nienhaus, G.U.; Bourgeois, D. Structural characterization of IrisFP, an optical highlighter undergoing multiple photo-induced transformations. Proc. Natl. Acad. Sci. USA 2008, 105, 18343–18348. [Google Scholar] [CrossRef] [Green Version]
- Cubitt, A.B.; Woollenweber, L.A.; Heim, R. Understanding structure-function relationships in the Aequorea victoria green fluorescent protein. Methods Cell Biol. 1999, 58, 19–30. [Google Scholar] [CrossRef]
- Pédelacq, J.-D.; Cabantous, S.; Tran, T.; Terwilliger, T.; Waldo, G. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2005, 24, 79–88. [Google Scholar] [CrossRef]
- Adam, V.; Nienhaus, K.; Bourgeois, D.; Nienhaus, G.U. Structural basis of enhanced photoconversion yield in green fluorescent protein-like protein Dendra2. Biochemistry 2009, 48, 4905–4915. [Google Scholar] [CrossRef]
- Macel, M.L.; Ristoratore, F.; Locascio, A.; Spagnuolo, A.; Sordino, P.; D’Aniello, S. Sea as a color palette: The ecology and evolution of fluorescence. Zool. Lett. 2020, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- Patterson, G.H.; Lippincott-Schwartz, J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science 2002, 297, 1873–1877. [Google Scholar] [CrossRef] [PubMed]
- McKinney, S.A.; Murphy, C.S.; Hazelwood, K.L.; Davidson, M.W.; Looger, L.L. A bright and photostable photoconvertible fluorescent protein. Nat. Methods. 2009, 6, 131–133. [Google Scholar] [CrossRef] [PubMed]
- Shaner, N.C.; Campbell, R.E.; Steinbach, P.A.; Giepmans, B.N.; Palmer, A.E.; Tsien, R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004, 22, 1567–1572. [Google Scholar] [CrossRef] [PubMed]
- Goulding, A.; Shrestha, S.; Dria, K.; Hunt, E.; Deo, S.K. Red fluorescent protein variants with incorporated non-natural amino acid analogues. Protein Eng. Des. Sel. 2008, 21, 101–106. [Google Scholar] [CrossRef] [Green Version]
- Kredel, S.; Oswald, F.; Nienhaus, K.; Deuschle, K.; Röcker, C.; Wolff, M.; Heilker, R.; Nienhaus, G.U.; Wiedenmann, J. mRuby, a bright monomeric red fluorescent protein for labeling of subcellular structures. PLoS ONE 2009, 4, 4391. [Google Scholar] [CrossRef]
- Subach, F.V.; Patterson, G.H.; Manley, S.; Gillette, J.M.; Lippincott-Schwartz, J.; Verkhusha, V.V. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat. Methods 2009, 6, 153–159. [Google Scholar] [CrossRef] [Green Version]
- Shaner, N.C.; Patterson, G.H.; Davidson, M.W. Advances in fluorescent protein technology. Cell Sci. 2007, 120, 4247–4260. [Google Scholar] [CrossRef] [Green Version]
- Campbell, R.E.; Tour, O.; Palmer, A.E.; Steinbach, P.A.; Zacharias, D.A.; Tsien, R.Y. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 2002, 99, 7877–7882. [Google Scholar] [CrossRef] [Green Version]
- Bulina, M.E.; Chudakov, D.M.; Britanova, O.V.; Yanushevich, Y.G.; Starorevor, D.B.; Chepurnykh, T.V.; Merlzlyak, E.M.; Shkrob, M.A.; Lukyanov, S.; Lukyanov, K.A. A genetically encoded photosensitizer. Nat. Biotechnol. 2006, 24, 95–99. [Google Scholar] [CrossRef]
- Shcherbo, D.; Merzlyak, E.M.; Chepurnykh, T.V.; Fradkov, A.F.; Ermakova, G.V.; Solovieva, E.A.; Lukyanov, K.A.; Bogdanova, E.A.; Zaraisky, A.G.; Lukyanov, S.; et al. Bright far-red fluorescent protein for whole-body imaging. Nat. Methods. 2007, 4, 741–746. [Google Scholar] [CrossRef] [PubMed]
- Fradkov, A.F.; Verkhusha, V.V.; Staroverov, D.B.; Bulina, M.E.; Yanushevich, Y.G.; Martynov, V.I.; Lukyanov, S.; Lukyanov, K.A. Far-red fluorescent tag for protein labelling. Biochem. J. 2002, 368, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Kredel, S.; Nienhaus, K.; Oswald, F.; Wolff, M.; Ivanchenko, S.; Cymer, F.; Jeromin, A.; Michel, F.J.; Spindler, K.D.; Heilker, R.; et al. Optimized and far-red-emitting variants of fluorescent protein eqFP611. Chem. Biol. 2008, 15, 224–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chudakov, D.M.; Belousov, V.V.; Zaraisky, A.G.; Novoselov, V.V.; Staroverov, D.B.; Zorov, D.B.; Lukyanov, S.; Lukyanov, K.A. Kindling fluorescent proteins for precise in vivo photolabeling. Nat. Biotechnol. 2003, 21, 191–194. [Google Scholar] [CrossRef]
- Shu, X.; Shaner, N.C.; Yarbrough, C.A.; Tsien, R.Y.; Remington, S.J. Novel chromophores and buried charges control color in mFruits. Biochemistry 2006, 45, 9639–9647. [Google Scholar] [CrossRef]
- Kikuchi, A.; Fukumura, E.; Karasawa, S.; Mizuno, H.; Miyawaki, A.; Shiro, Y. Structural characterization of a thiazoline-containing chromophore in an orange fluorescent protein, monomeric Kusabira Orange. Biochemistry 2008, 47, 11573–11580. [Google Scholar] [CrossRef]
- Hasegawa, J.; Ise, T.; Fujimoto, K.J.; Kikuchi, A.; Fukumura, E.; Miyawaki, A.; Shiro, Y. Excited states of fluorescent proteins, mKO and DsRed: Chromophore−protein electrostatic interaction behind the color variations. J. Phys. Chem. B 2010, 114, 2971–2979. [Google Scholar] [CrossRef]
- Bloemberg, G.V. Microscopic analysis of plant-bacterium interactions using auto fluorescent proteins. Eur. J. Plant Pathol. 2007, 119, 301–309. [Google Scholar] [CrossRef] [Green Version]
- Bloemberg, G.V.; Lugtenberg, B.J.J. Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr. Opin. Plant Biol. 2001, 4, 343–350. [Google Scholar] [CrossRef]
- Lübeck, P.S.; Hansen, M.; Sørensen, J. Simultaneous detection of the establishment of seed-inoculated Pseudomonas fluorescens strain DR54 and native soil bacteria on sugar beet root surfaces using fluorescence antibody and in situ hybridization techniques. FEMS Microbiol. Ecol. 2000, 33, 11–19. [Google Scholar] [CrossRef]
- Normander, B.; Hendriksen, N.B.; Nybroe, O. Green fluorescent protein-marked Pseudomonas fluorescens: Localization, viability, and activity in the natural barley rhizosphere. Appl. Environ. Microbiol. 1999, 65, 4646–4651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rochat, L.; Péchy-Tarr, M.; Baehler, E.; Maurhofer, M.; Keel, C. Combination of fluorescent reporters for simultaneous monitoring of root colonization and antifungal gene expression by a biocontrol pseudomonad on cereals with flow cytometry. Mol. Plant Microbe Interact. 2010, 23, 949–961. [Google Scholar] [CrossRef] [Green Version]
- Gage, D.J.; Bobo, T.; Long, S.R. Use of green fluorescent protein to visualize the early events of symbiosis between Rhizobium meliloti and alfalfa (Medicago sativa). J. Bacteriol. 1996, 178, 7159–7166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bloemberg, G.V.; O’Toole, G.A.; Lugtenberg, B.J.; Kolter, R. Green fluorescent protein as a marker for Pseudomonas spp. Appl. Environ. Microbiol. 1997, 63, 4543–4551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tombolini, R.; Unge, A.; Davey, M.E.; de Bruijn, F.J.; Jansson, J.K. Flow cytometric and microscopic analysis of GFP-tagged Pseudomonas fluorescens bacteria. FEMS Microbiol. Ecol. 1997, 22, 17–28. [Google Scholar] [CrossRef]
- Lanna Filho, R.; de Souza, R.M.; Ferreira, A.; Quecine, M.C.; Alves, E.; de Azevedo, J.L. Biocontrol activity of Bacillus against a GFP-marked Pseudomonas syringae pv. tomato on tomato phylloplane. Australas. Plant Pathol. 2013, 42, 643–651. [Google Scholar] [CrossRef] [Green Version]
- Zavaleta-Mancera, H.; Valencia-Botín, A.; Mendoza-Onofre, L.; Silva-Rojas, H.; Valadez-Moctezuma, E. Use of green fluorescent protein to monitor the colonization of Pseudomonas syringae subsp. syringae on wheat seeds. Microsc. Microanal. 2007, 13, 298–299. [Google Scholar] [CrossRef]
- Akimoto-Tomiyama, C.; Furutani, A.; Ochiai, H. Real time live imaging of phytopathogenic bacteria Xanthomonas campestris pv. campestris MAFF106712 in ‘plant sweet home’. PLoS ONE 2014, 9, e94386. [Google Scholar] [CrossRef]
- So, J.-S.; Lim, H.T.; Oh, E.-T.; Heo, T.-R.; Koh, S.-C.; Leung, K.T.; Lee, H.; Trevors, J.T. Visualizing the infection process of Xanthomonas campestris in cabbage using green fluorescent protein. World J. Microbiol. Biotechnol. 2002, 18, 17–21. [Google Scholar] [CrossRef]
- Van der Wolf, J.; Kastelein, P.; da Silva Júnior, T.A.F.; Lelis, F.V.; van der Zouwen, P. Colonization of siliques and seeds of rapid cycling Brassica oleracea plants by Xanthomonas campestris pv. campestris after spray-inoculation of flower clusters. Eur. J. Plant Pathol. 2019, 154, 445–461. [Google Scholar] [CrossRef] [Green Version]
- Fujie, M.; Takamoto, H.; Kawasaki, T.; Fujiwara, A.; Yamada, T. Monitoring growth and movement of Ralstonia solanacearum cells harboring plasmid pRSS12 derived from bacteriophage ϕRSS1. J. Biosci. Bioeng. 2010, 109, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Satsuma, H.; Fujie, M.; Usami, S.; Yamada, T. Monitoring of phytopathogenic Ralstonia solanacearum cells using green fluorescent protein-expressing plasmid derived from bacteriophage ϕRSS1. J. Biosci. Bioeng. 2007, 104, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, Y.; Cai, L.; Zhang, H.; Shi, J.; Yuan, Y. Factors affecting the virulence of Ralstonia solanacearum and its colonization on tobacco roots. Plant Pathol. 2017, 66, 1345–1356. [Google Scholar] [CrossRef]
- Chalupowicz, L.; Zellermann, E.-M.; Fluegel, M.; Dror, O.; Eichenlaub, R.; Gartemann, K.-H.; Savidor, A.; Sessa, G.; Iraki, N.; Barash, I.; et al. Colonization and movement of GFP-labeled Clavibacter michiganensis subsp. michiganensis during tomato infection. Phytopathology 2012, 102, 23–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lelis, F.M.V.; Czajkowski, R.; de Souza, R.M.; Ribeiro, D.H.; van der Wolf, J.M. Studies on the colonization of axenically grown tomato plants by a GFP-tagged strain of Clavibacter michiganensis subsp. michiganensis. Eur. J. Plant Pathol. 2014, 139, 53–66. [Google Scholar] [CrossRef]
- Czajkowski, R.; de Boer, W.J.; van Veen, J.A.; van der Wolf, J.M. Downward vascular translocation of a green fluorescent protein-tagged strain of Dickeya sp. (biovar 3) from stem and leaf inoculation sites on potato. Phytopathology 2010, 100, 1128–1137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czajkowski, R.; de Boer, W.J.; Velvis, H.; van der Wolf, J.M. Systemic colonization of potato plants by a soilborne, green fluorescent protein-tagged strain of Dickeya sp. biovar 3. Phytopathology 2010, 100, 134–142. [Google Scholar] [CrossRef] [Green Version]
- Czajkowski, R.; de Boer, W.J.; van Veen, J.A.; van der Wolf, J.M. Characterization of bacterial isolates from rotting potato tuber tissue showing antagonism to Dickeya sp. biovar 3 in vitro and in planta. Plant Pathol. 2012, 61, 169–182. [Google Scholar] [CrossRef]
- Czajkowski, R.; de Boer, W.J.; van der Zouwen, P.S.; Kastelein, P.; Jafra, S.; de Haan, E.G.; van den Bovenkamp, G.W.; van der Wolf, J.M. Virulence of ‘Dickeya solani’ and Dickeya dianthicola biovar-1 and -7 strains on potato (Solanum tuberosum). Plant Pathol. 2013, 62, 597–610. [Google Scholar] [CrossRef]
- Kubheka, G.C.; Coutinho, T.; Moleleki, N.; Moleleki, L.N. Colonisation patterns of a mCherry-tagged Pectobacterium carotovorum subsp. brasiliense strain in potato plants. Phytopathology 2013, 103, 1268–1279. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Yuan, L.; Shi, Y.; Xie, X.; Chai, A.; Wang, Q.; Li, B. Comparative genomic analysis of Pectobacterium carotovorum subsp. brasiliense SX309 provides novel insights into its genetic and phenotypic features. BMC Genom. 2019, 20, 486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chane, A.; Barbey, C.; Robert, M.; Merieau, A.; Konto-Ghiorghi, Y.; Beury-Cirou, A.; Feuilloley, M.; Pátek, M.; Gobert, V.; Latour, X. Biocontrol of soft-rot: Confocal microscopy highlights virulent pectobacterial communication and its jamming by rhodococcal quorum-quenching. Mol. Plant Microbe Interact. 2019, 32, 802–812. [Google Scholar] [CrossRef] [PubMed]
- Kastelein, P.; Förch, M.G.; Krijger, M.C.; van der Zouwen, P.S.; van den Berg, W.; van der Wolf, J.M. Systemic colonization of potato plants resulting from potato haulm inoculation with Dickeya solani or Pectobacterium parmentieri. Can. J. Plant Pathol. 2020, in press. [Google Scholar] [CrossRef]
- Narváez-Barragán, D.A.; De Sandozequi, A.; Rodríguez, M.; Estrada, K.; Tovar-Herrera, O.E.; Martínez-Anaya, C. Analysis of two Mexican Pectobacterium brasiliense strains reveals an inverted relationship between c-di-GMP levels with exopolysaccharide production and swarming motility. Microbiol. Res. 2020, 235, 126427. [Google Scholar] [CrossRef] [PubMed]
- Golan, A.; Kerem, Z.; Tun, O.M.; Luzzatto, T.; Lipsky, A.; Yedidia, I. Combining flow cytometry and gfp reporter gene for quantitative evaluation of Pectobacterium carotovorum ssp. carotovorum in Ornithogalum dubium plantlets. J. Appl. Microbiol. 2010, 108, 1136–1144. [Google Scholar] [CrossRef]
- Fikowicz-Krosko, J.; Czajkowski, R. Systemic colonisation and expression of disease symptoms on bittersweet nightshade (Solanum dulcamara L.) infected with a GFP-tagged Dickeya solani IPO2222 (IPO2254). Plant Dis. 2018, 102, 619–627. [Google Scholar] [CrossRef] [Green Version]
- Czajkowski, R.; van der Wolf, J.M.; Krolicka, A.; Ozymko, Z.; Narajczyk, M.; Kaczynska, N.; Lojkowska, E. Salicylic acid can reduce infection symptoms caused by Dickeya solani in tissue culture grown potato (Solanum tuberosum L.) plants. Eur. J. Plant Pathol. 2015, 141, 545–558. [Google Scholar] [CrossRef] [Green Version]
- Tamayo, R.; Pratt, J.T.; Camilli, A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 2007, 61, 131–148. [Google Scholar] [CrossRef] [Green Version]
- Diallo, S.; Crépin, A.; Barbey, C.; Orange, N.; Burini, J.-F.; Latour, X. Mechanisms and recent advances in biological control mediated through the potato rhizosphere. FEMS Microbiol. Ecol. 2011, 75, 351–364. [Google Scholar] [CrossRef]
- Kloepper, J.W. Effect of seed piece inoculation with plant growth-promoting rhizobacteria on population of Erwinia carotovora on potato roots and in daughter tubers. Phytopathology 1983, 73, 217–219. [Google Scholar] [CrossRef] [Green Version]
- Compant, S.; Clément, C.; Sessitsch, A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 2010, 42, 669–678. [Google Scholar] [CrossRef] [Green Version]
- Czajkowski, R.; Krzyżanowska, D.; Karczewska, J.; Atkinson, S.; Przysowa, J.; Lojkowska, E.; Williams, P.; Jafra, S. Inactivation of AHLs by Ochrobactrum sp. A44 depends on the activity of a novel class of AHL acylase. Environ. Microbiol. Rep. 2011, 3, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Crépin, A.; Barbey, C.; Cirou, A.; Tannières, M.; Orange, N.; Feuilloley, M.; Dessaux, Y.; Burini, J.-F.; Faure, D.; Latour, X. Biological control of pathogen communication in the rhizosphere: A novel approach applied to potato soft rot due to Pectobacterium atrosepticum. Plant Soil 2012, 358, 27–37. [Google Scholar] [CrossRef]
- Gerayeli, N.; Baghaee-Ravari, S.; Tarighi, S. Evaluation of the antagonistic potential of Bacillus strains against Pectobacterium carotovorum subsp. carotovorum and their role in the induction of resistance to potato soft rot infection. Eur. J. Plant Pathol. 2018, 150, 1049–1063. [Google Scholar] [CrossRef]
- Krzyżanowska, D.M.; Ossowicki, A.; Rajewska, M.; Maciąg, T.; Jabłońska, M.; Obuchowski, M.; Heeb, S.; Jafra, S. When genome-based approach meets the ‘old but good’: Revealing genes involved in the antibacterial activity of Pseudomonas sp. P482 against soft rot pathogens. Front. Microbiol. 2016, 7, 782. [Google Scholar] [CrossRef]
- Raoul des Essarts, Y.; Cigna, J.; Quêtu-Laurent, A.; Caron, A.; Munier, E.; Beury-Cirou, A.; Hélias, V.; Faure, D. Biocontrol of the potato blackleg and soft rot diseases caused by Dickeya dianthicola. Appl. Environ. Microbiol. 2015, 82, 268–278. [Google Scholar] [CrossRef] [Green Version]
- Tsuda, K.; Tsuji, G.; Higashiyama, M.; Ogiyama, H.; Umemura, K.; Mitomi, M.; Kubo, Y.; Kosaka, Y. Biological control of bacterial soft rot in Chinese cabbage by Lactobacillus plantarum strain BY under field conditions. Biol. Control 2016, 100, 63–69. [Google Scholar] [CrossRef]
- Li, Z.; Wang, T.; Luo, X.; Li, X.; Xia, C.; Zhao, Y.; Ye, X.; Huang, Y.; Gu, X.; Cao, H.; et al. Biocontrol potential of Myxococcus sp. strain BS against bacterial soft rot of calla lily caused by Pectobacterium carotovorum. Biol. Control 2018, 126, 36–44. [Google Scholar] [CrossRef]
- Cui, W.; He, P.; Munir, S.; He, P.; He, Y.; Li, X.; Yang, L.; Wang, B.; Wu, Y.; He, P. Biocontrol of soft rot of Chinese cabbage using an endophytic bacterial strain. Front Microbiol. 2019, 10, 1471. [Google Scholar] [CrossRef] [Green Version]
- Barbey, C.; Crépin, A.; Bergeau, D.; Ouchiha, A.; Mijouin, L.; Taupin, L.; Orange, N.; Feuilloley, M.; Dufour, A.; Burini, J.F.; et al. In planta biocontrol of Pectobacterium atrosepticum by Rhodococcus erythropolis involves silencing of pathogen communication by the rhodococcal gamma-lactone catabolic pathway. PLoS ONE 2013, 8, e66642. [Google Scholar] [CrossRef]
- Garge, S.S.; Nerurkar, A.S. Evaluation of quorum quenching Bacillus spp. for their biocontrol traits against Pectobacterium carotovorum subsp. carotovorum causing soft rot. Biocat. Agric. Biotechnol. 2017, 9, 48–57. [Google Scholar] [CrossRef]
- Krzyżanowska, D.M.; Potrykus, M.; Golanowska, M.; Polonis, K.; Gwizdek-Wisniewska, A.; Lojkowska, E.; Jafra, S. Rhizosphere bacteria as potential biocontrol agents against soft rot caused by various Pectobacterium and Dickeya spp. strains. J. Plant Pathol. 2012, 94, 367–378. [Google Scholar] [CrossRef]
- Jafra, S.; Przysowa, J.; Gwizdek-Wiśniewska, A.; van der Wolf, J.M. Potential of bulb-associated bacteria for biocontrol of hyacinth soft rot caused by Dickeya zeae. J. Appl. Microbiol. 2009, 106, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Czajkowski, R.; de Boer, W.J.; van Veen, J.A.; van der Wolf, J.M. Studies on the interaction between the biocontrol agent, Serratia plymuthica A30, and blackleg-causing Dickeya sp. (biovar 3) in potato (Solanum tuberosum): Interaction between Serratia A30 and Dickeya sp. IPO2222. Plant Pathol. 2012, 61, 677–688. [Google Scholar] [CrossRef]
- Maciag, T.; Krzyzanowska, D.M.; Jafra, S.; Siwinska, J.; Czajkowski, R. The great five-An artificial bacterial consortium with antagonistic activity towards Pectobacterium spp. and Dickeya spp.: Formulation, shelf life, and the ability to prevent soft rot of potato in storage. Appl. Microbiol. Biotechnol. 2020, 104, 4547–4561. [Google Scholar] [CrossRef] [Green Version]
- Hadizadeh, I.; Peivastegan, B.; Hannukkala, A.; van der Wolf, J.M.; Nissinen, R.; Pirhonen, M. Biological control of potato soft rot caused by Dickeya solani and the survival of bacterial antagonists under cold storage conditions. Plant Pathol. 2019, 68, 297–311. [Google Scholar] [CrossRef] [Green Version]
- Krzyżanowska, D.M.; Obuchowski, M.; Bikowski, M.; Rychłowski, M.; Jafra, S. Colonization of potato rhizosphere by GFP-tagged Bacillus subtilis MB73/2, Pseudomonas sp. P482 and Ochrobactrum sp. A44 shown on large sections of roots using enrichment sample preparation and confocal laser scanning microscopy. Sensors 2012, 12, 17608–17619. [Google Scholar] [CrossRef]
- Krzyżanowska, D.M.; Maciąg, T.; Ossowicki, A.; Rajewska, M.; Kaczyński, Z.; Czerwicka, M.; Rąbalski, Ł.; Czaplewska, P.; Jafra, S. Ochrobactrum quorumnocens sp. nov., a quorum quenching bacterium from the potato rhizosphere, and comparative genome analysis with related type strains. PLoS ONE 2019, 14, e0210874. [Google Scholar] [CrossRef]
- Jafra, S.; Przysowa, J.; Czajkowski, R.; Michta, A.; Garbeva, P.; van der Wolf, J. Detection and characterization of bacteria from the potato rhizosphere degrading N-acyl-homoserine lactone. Can. J. Microbiol. 2006, 52, 1006–1015. [Google Scholar] [CrossRef]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal-response systems in gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef]
- Pierson, E.A.; Wood, D.W.; Cannon, J.A.; Blachere, F.M.; Pierson, L.S., 3rd. Interpopulation signaling via N-acyl-homoserine lactones among bacteria in the wheat rhizosphere. Mol. Plant Microbe Interact. 1998, 11, 1078–1084. [Google Scholar] [CrossRef] [Green Version]
- Morello, J.E.; Pierson, E.A.; Pierson, L.S., 3rd. Negative cross-communication among wheat rhizosphere bacteria: Effect on antibiotic production by the biological control bacterium Pseudomonas aureofaciens 30-84. Appl. Environ. Microbiol. 2004, 70, 3103–3109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charlesworth, J.; Kimyon, O.; Manefield, M.; Burns, B.P. Detection and characterization of N-acyl-l-homoserine lactones using GFP-based biosensors in conjunction with thin-layer chromatography. J. Microbiol. Methods 2015, 118, 164–167. [Google Scholar] [CrossRef] [PubMed]
- Christensen, L.D.; van Gennip, M.; Jakobsen, T.H.; Givskov, M.; Bjarnsholt, T. Imaging N-acyl homoserine lactone quorum sensing in vivo. Methods Mol. Biol. 2011, 692, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Riedel, K.; Hentzer, M.; Geisenberger, O.; Huber, B.; Steidle, A.; Wu, H.; Høiby, N.; Givskov, M.; Molin, S.; Eberl, L. N-acylhomoserine- lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 2001, 147, 3249–3262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gantner, S.; Schmid, M.; Dürr, C.; Schuhegger, R.; Steidle, A.; Hutzler, P.; Langebartels, C.; Eberl, L.; Hartmann, A.; Dazzo, F.B. In situ quantitation of the spatial scale of calling distances and population density-independent N-acylhomoserine lactone-mediated communication by rhizobacteria colonized on plant roots. FEMS Microbiol. Ecol. 2006, 56, 188–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Coulthurst, S.J.; Pritchard, L.; Hedley, P.E.; Ravensdale, M.; Humphris, S.; Burr, T.; Takle, G.; Brurberg, M.-B.; Birch, P.R.J.; et al. Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum. PLoS Pathog. 2008, 20, e1000093. [Google Scholar] [CrossRef]
- Chane, A.; Barbey, C.; Bourigault, Y.; Beury-Cirou, A.; Bouteiller, M.; Merieau, A.; Konto-Ghiorghi, Y.; Feuilloley, M.; Gobert, V.; Latour, X. Informational war as therapy to silence blackleg and soft-rot diseases. J. Plant Pathol. 2019, 101, 869. [Google Scholar] [CrossRef]
- Chane, A.; Bourigault, Y.; Bouteiller, M.; Konto-Ghiorghi, Y.; Merieau, A.; Barbey, C.; Latour, X. Close-up on a bacterial informational war in the geocaulosphere. Can. J. Microbiol. 2020, 66, 447–454. [Google Scholar] [CrossRef]
- Dong, Y.H.; Wang, L.H.; Xu, J.L.; Zhang, H.B.; Zhang, X.F.; Zhang, L.H. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 2001, 411, 813–817. [Google Scholar] [CrossRef]
- Chane, A.; Barbey, C.; Bourigault, Y.; Maillot, O.; Rodrigues, S.; Bouteiller, M.; Merieau, A.; Konto-Ghiorghi, Y.; Beury-Cirou, A.; Gattin, R.; et al. A flavor lactone mimicking AHL quorum-sensing signals exploits the broad affinity of the QsdR regulator to stimulate transcription of the rhodococcal qsd operon involved in quorum-quenching and biocontrol activities. Front. Microbiol. 2019, 10, 786. [Google Scholar] [CrossRef] [PubMed]
- Grandclément, C.; Tannières, M.; Moréra, S.; Dessaux, Y.; Faure, D. Quorum quenching: Role in nature and applied developments. FEMS Microbiol. Rev. 2016, 40, 86–116. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.H.; Wang, L.H.; Zhang, L.H. Quorum-quenching microbial infections: Mechanisms and implications. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2007, 362, 1201–1211. [Google Scholar] [CrossRef] [PubMed]
- Bergonzi, C.; Schwab, M.; Naik, T.; Elias, M. The structural determinants accounting for the broad substrate specificity of the quorum quenching lactonase GcL. Chembiochem 2019, 20, 1848–1855. [Google Scholar] [CrossRef] [PubMed]
- Schwab, M.; Bergonzi, C.; Sakkos, J.; Staley, C.; Zhang, Q.; Sadowsky, M.J.; Aksan, A.; Elias, M. Signal disruption leads to changes in bacterial community population. Front. Microbiol. 2019, 10, 611. [Google Scholar] [CrossRef]
- Vesuna, A.; Nerurkar, A.S. Enzymatic quorum quenching for virulence attenuation of phytopathogenic bacteria. In Biotechnological Applications of Quorum Sensing Inhibitors; Kalia, V., Ed.; Springer: Singapore, Singapore, 2018; pp. 447–473. [Google Scholar] [CrossRef]
- Vesuna, A.P.; Nerurkar, A.S. Biocontrol impact of AHL degrading actinobacteria on quorum sensing regulated virulence of phytopathogen Pectobacterium carotovorum subsp. carotovorum BR1. Plant Soil 2020, 453, 371–388. [Google Scholar] [CrossRef]
- Cirou, A.; Raffoux, A.; Diallo, S.; Latour, X.; Dessaux, Y.; Faure, D. Gamma-caprolactone stimulates growth of quorum-quenching Rhodococcus populations in a large-scale hydroponic system for culturing Solanum tuberosum. Res. Microbiol. 2011, 162, 945–950. [Google Scholar] [CrossRef]
- Cirou, A.; Mondy, S.; An, S.; Charrier, A.; Sarrazin, A.; Thoison, O.; DuBow, M.; Faure, D. Efficient biostimulation of native and introduced quorum-quenching Rhodococcus erythropolis populations is revealed by a combination of analytical chemistry, microbiology, and pyrosequencing. Appl. Environ. Microbiol. 2012, 78, 481–492. [Google Scholar] [CrossRef] [Green Version]
- Barbey, C.; Chane, A.; Burini, J.-F.; Maillot, O.; Merieau, A.; Gallique, M.; Beury-Cirou, A.; Konto-Ghiorghi, Y.; Feuilloley, M.; Gobert, V.; et al. A rhodococcal transcriptional regulatory mechanism detects the common lactone ring of AHL quorum-sensing signals and triggers the quorum-quenching response. Front. Microbiol. 2018, 9, 2800. [Google Scholar] [CrossRef]
- Latour, X.; Barbey, C.; Chane, A.; Groboillot, A.; Burini, J.-F. Rhodococcus erythropolis and its γ-lactone catabolic pathway: An unusual rhodococcal quorum-quenching pathway regulation biocontrol system that disrupts pathogen quorum sensing communication. Agronomy 2013, 3, 816–838. [Google Scholar] [CrossRef] [Green Version]
- El Sahili, A.; Kwasiborski, A.; Mothe, N.; Velours, C.; Legrand, P.; Moréra, S.; Faure, D. Natural guided genome engineering reveals transcriptional regulators controlling quorum-sensing signal degradation. PLoS ONE 2015, 10, e0141718. [Google Scholar] [CrossRef] [PubMed]
- Knoppová, M.; Phensaijai, M.; Veseý, M.; Zemanová, M.; Nešvera, J.; Pátek, M. Plasmid vectors for testing in vivo promoter activities in Corynebacterium glutamicum and Rhodococcus erythropolis. Curr. Microbiol. 2007, 55, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Elowitz, M.B.; Levine, A.J.; Siggia, E.D.; Swain, P.S. Stochastic gene expression in a single cell. Science 2002, 297, 1183–1186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gangwe Nana, G.Y.; Ripoll, C.; Cabin-Flaman, A.; Gibouin, D.; Delaune, A.; Janniere, L.; Grancher, G.; Chagny, G.; Loutelier-Bourhis, C.; Lentzen, E.; et al. Division-based, growth rate diversity in bacteria. Front. Microbiol. 2018, 9, 849. [Google Scholar] [CrossRef] [PubMed]
- Plamont, M.A.; Billon-Denis, E.; Maurin, S.; Gauron, C.; Pimenta, F.M.; Specht, C.G.; Shi, J.; Quérard, J.; Pan, B.; Rossignol, J.; et al. Small fluorescence-activating and absorption-shifting tag for tunable protein imaging in vivo. Proc. Natl. Acad. Sci. USA 2016, 113, 497–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pena, M.M.; Teper, D.; Ferreira, H.; Wang, N.; Sato, K.U.; Ferro, M.I.T.; Ferro, J.A. mCherry fusions enable the subcellular localization of periplasmic and cytoplasmic proteins in Xanthomonas sp. PLoS ONE 2020, 15, e0236185. [Google Scholar] [CrossRef] [PubMed]



| Name | Excitation/Emission Used (nm) | Carrier Strain | Plasmid Probe | Application | Reference |
|---|---|---|---|---|---|
| GFP | 485/505 | D. solani IPO2254 (bv.3) | pPROBE-AT-gfp | Localization, spread | [97,98,99] |
| GFP | 495/505 | D. solani IPO2253 (bv.1) | pPROBE-AT-gfp | Localization, spread | [100] |
| DsRed | 532/610 | D. solani IPO3012 (bv.3) | pRZ-T3-dsred | Localization, spread | [100] |
| DsRed | 557/579 | D. dianthicola IPO3018 (bv.7) | pRZ-T3-dsred | Localization, spread | [100] |
| GFP | 495/505 | D. dianthicola IPO3019 (bv.7) | pPROBE-AT-gfp | Localization, spread | [100] |
| mCherry | 560/615 | P. brasiliense 1962 | pMP7604-mCherry | Localization | [101] |
| Egfp | 488/507 | P. brasiliense 1962 | pMP4657-egfp | Localization | [29] |
| Egfp | 488/507 | P. brasiliense 1962ΔexpI 1 | pMP4657-egfp | Localization | [29] |
| GFP | 488/509 | P. brasiliense SX309 | pSMC21-gfp | Localization | [102] |
| CFP and GFPASV | 405/477488/509 | P. atrosepticum CFBP 6276 | pME6000-luxR-PluxI::gfpASV-cfp | Localization and endogenous QS detection 2 | [103] |
| CFP and GFPASV | 405/477488/509 | P. atrosepticum CFBP 6276-EI 1 | pME6000-luxR-PluxI::gfpASV-cfp | Localization and exogenous QS detection 2 | [103] |
| GFP | 495/505 | P. parmentieri IPO3399 | pPROBE-AT-gfp | Localization, spread | [104] |
| AmCyan and TurboRFP | 420/450550/580 | P. atrosepticum SCRI1043 | pFY435 3 | c-di-GMP production | [105] |
| AmCyan and TurboRFP | 420/450550/580 | P. brasiliense BF20 | pFY435 3 | c-di-GMP production | [105] |
| AmCyan and TurboRFP | 420/450 550/580 | P. brasiliense BF45 | pFY435 3 | c-di-GMP production | [105] |
| Process | Bacterial Strain | Host Plant | Fluorescent Protein Used | Reference |
|---|---|---|---|---|
| Bidirectional bacterial movement inside plants | D. solani strain IPO2254 P. carotovorum subsp. carotovorum strains Pcc3 and Pcc13 | Potato (Solanum tuberosum L.) Sun star (Ornithogalum dubium) | GFP GFP | [97,98,106] |
| Symptomless dissemination of bacteria inside plants | D. solani strain IPO2254 | Potato (Solanum tuberosum L.) | GFP | [97,98] |
| Colonization of (progeny) tubers | D. solani strain IPO2254 | Potato (Solanum tuberosum L.) | GFP | [97,98] |
| Presence and spread of bacteria inside tolerant vs. susceptible host | P. brasiliense strain Pcb1692 | Potato (Solanum tuberosum L.) | mCherry | [101] |
| Fate of bacterial strains in salicylic treated host | D. solani strain IPO2254 | Potato (Solanum tuberosum L.) | GFP | [108] |
| Fate of AHL-deficient bacterial variants in host plant | P. brasiliense strain Pcb1692 | Potato (Solanum tuberosum L.) | GFP | [29] |
| Systemic colonization of alternative plant hosts | D. solani strain IPO2254 | Bittersweet nightshade (Solanum dulcamara L.) | GFP | [107] |
| Colonization of plants after spray-inoculation | D. solani strain IPO2254 P. parmentieri strain IPO3399 | Potato (Solanum tuberosum L.) | GFP | [104] |
| Detection of quorum-sensing activity | P. atrosepticum strain 6276 | Potato (Solanum tuberosum L.) | GFP and CFP | [103] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bourigault, Y.; Chane, A.; Barbey, C.; Jafra, S.; Czajkowski, R.; Latour, X. Biosensors Used for Epifluorescence and Confocal Laser Scanning Microscopies to Study Dickeya and Pectobacterium Virulence and Biocontrol. Microorganisms 2021, 9, 295. https://doi.org/10.3390/microorganisms9020295
Bourigault Y, Chane A, Barbey C, Jafra S, Czajkowski R, Latour X. Biosensors Used for Epifluorescence and Confocal Laser Scanning Microscopies to Study Dickeya and Pectobacterium Virulence and Biocontrol. Microorganisms. 2021; 9(2):295. https://doi.org/10.3390/microorganisms9020295
Chicago/Turabian StyleBourigault, Yvann, Andrea Chane, Corinne Barbey, Sylwia Jafra, Robert Czajkowski, and Xavier Latour. 2021. "Biosensors Used for Epifluorescence and Confocal Laser Scanning Microscopies to Study Dickeya and Pectobacterium Virulence and Biocontrol" Microorganisms 9, no. 2: 295. https://doi.org/10.3390/microorganisms9020295
APA StyleBourigault, Y., Chane, A., Barbey, C., Jafra, S., Czajkowski, R., & Latour, X. (2021). Biosensors Used for Epifluorescence and Confocal Laser Scanning Microscopies to Study Dickeya and Pectobacterium Virulence and Biocontrol. Microorganisms, 9(2), 295. https://doi.org/10.3390/microorganisms9020295