PICCOLO White-Light and Narrow-Band Imaging Colonoscopic Dataset: A Performance Comparative of Models and Datasets
Abstract
:Featured Application
Abstract
1. Introduction
2. Materials and Methods
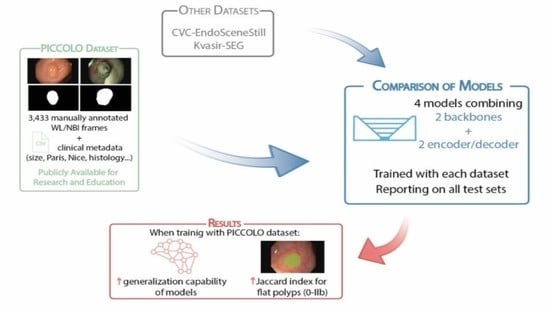
2.1. PICCOLO Dataset
2.1.1. Acquisition Protocol
- Patients included in the colon cancer screening and surveillance program were informed about the study and asked for permission to collect images/videos obtained during routine colonoscopy with the associated information via informed consent and patient information sheet. If the patient gave permission, the rest of the protocol was followed.
- If a suspicious lesion was found during the procedure, it was resected using the most appropriate method and sent to the Department of Pathological Anatomy for diagnosis.
- Images/videos with the associated clinical information were anonymized.
- Videos were analysed and edited/processed by the gastroenterologists who performed the colonoscopy. The video of the full procedure was divided into fragments, indicating if the tissue was healthy or pathological and the region on the colon where it was found. Further details on the annotation process are given in the following subsection.
- The gastroenterologist completed part of the associated metadata:
- The pathologist completed part of the associated metadata:
- Final diagnosis;
- Histological stratification.
- Clinical metadata were exported into a csv file.
2.1.2. Annotation Protocol
- Video clips were processed to extract the individual frames. Uninformative frames were discarded from this process and they includeD (Figure S1):
- Frames outside the patient;
- Blurry frames;
- Frames with high presence of bubbles;
- Frames with high presence of stool;
- Transition frames between WL and NBI.
- All frames were analysed and categorized into showing a polyp or not, as well as identifying the type of light source (WL or NBI).
- One out of 25 polyp frames (i.e., one frame per second) was selected to be manually annotated.
- A researcher prepared three equally distributed sets of images to be processed using GTCreator [21]. Each set was manually annotated by one independent expert gastroenterologist with more than 15,000–25,000 (Á.J.C.; F.P.) and 500 (B.G.) colonoscopies. Furthermore, a void mask was also generated to indicate the valid endoscopic area of the image.
- Segmented frames were collected and revised by a researcher to check completeness of the dataset prior to its use.
- Manually segmented masks were automatically corrected with the void mask to adjust segmentations to the endoscopic image area.
2.1.3. PICCOLO Dataset Details
- -
- Frame itself: png files showing the WL or NBI image.
- -
- Mask: Binary mask indicating the area corresponding to the lesion.
- -
- Void: Binary mask indicating the black area of the image.
2.2. Public Datasets
- CVC-EndoSceneStill [23]. It contains 912 WL images which are manually segmented. A distribution into training, validation and test sets is provided by the owners. Each set contains 547, 183 and 182 images, respectively. This dataset is available at http://www.cvc.uab.es/CVC-Colon/index.php/databases/cvc-endoscenestill/.
- Kvasir-SEG [24]. It contains 1000 WL images which are manually segmented. Distribution into training, validation and test sets has been randomly done, so each set includes 600, 200 and 200 images, respectively. This dataset is available at https://datasets.simula.no/kvasir-seg/.
2.3. Architectures and Training Process
2.4. Reporting
3. Results and Discussion
3.1. Characterization of the Datasets
3.2. Comparison of Models Performance
3.3. Influence of the Polyp Morphology in the Results
3.4. Current Limitations and Future Work
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- International Agency for Research on Cancer. Colorectal Cancer Factsheet; International Agency for Research on Cancer: Lyon, France, 2018. [Google Scholar]
- World Health Organization. World Cancer Report 2014; Stewart, B.W., Wild, C.P., Eds.; International Agency for Research on Cancer: Lyon, France, 2014; ISBN 978-92-832-0429-9. [Google Scholar]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2015. CA Cancer J. Clin. 2015, 65, 29. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Wiegering, A.; Ackermann, S.; Riegel, J.; Dietz, U.A.; Götze, O.; Germer, C.T.; Klein, I. Improved survival of patients with colon cancer detected by screening colonoscopy. Int. J. Colorectal Dis. 2016, 31, 1039–1045. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, M.F.; Thomas-Gibson, S.; Bugajski, M.; Bretthauer, M.; Rees, C.J.; Dekker, E.; Hoff, G.; Jover, R.; Suchanek, S.; Ferlitsch, M.; et al. Performance measures for lower gastrointestinal endoscopy: A European Society of Gastrointestinal Endoscopy (ESGE) quality improvement initiative. United Eur. Gastroenterol. J. 2017, 5, 309–334. [Google Scholar] [CrossRef]
- Lund, M.; Trads, M.; Njor, S.H.; Erichsen, R.; Andersen, B. Quality indicators for screening colonoscopy and colonoscopist performance and the subsequent risk of interval colorectal cancer: A systematic review. JBI Database Syst. Rev. Implement. Reports 2019. [Google Scholar] [CrossRef]
- Kim, N.H.; Jung, Y.S.; Jeong, W.S.; Yang, H.-J.; Park, S.-K.; Choi, K.; Park, D. Il Miss rate of colorectal neoplastic polyps and risk factors for missed polyps in consecutive colonoscopies. Intest. Res. 2017, 15, 411. [Google Scholar] [CrossRef] [Green Version]
- Klare, P.; Sander, C.; Prinzen, M.; Haller, B.; Nowack, S.; Abdelhafez, M.; Poszler, A.; Brown, H.; Wilhelm, D.; Schmid, R.M.; et al. Automated polyp detection in the colorectum: A prospective study (with videos). Gastrointest. Endosc. 2019, 89, 576–582. [Google Scholar] [CrossRef]
- Ishaq, S.; Siau, K.; Harrison, E.; Tontini, G.E.; Hoffman, A.; Gross, S.; Kiesslich, R.; Neumann, H. Technological advances for improving adenoma detection rates: The changing face of colonoscopy. Dig. Liver Dis. 2017, 49, 721–727. [Google Scholar] [CrossRef]
- Byrne, M.F.; Shahidi, N.; Rex, D.K. Will Computer-Aided Detection and Diagnosis Revolutionize Colonoscopy? Gastroenterology 2017, 153, 1460–1464. [Google Scholar] [CrossRef] [Green Version]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Kim, J.; Hong, J.; Park, H. Prospects of deep learning for medical imaging. Precis. Futur. Med. 2018, 2, 37–52. [Google Scholar] [CrossRef]
- Shen, D.; Wu, G.; Suk, H.-I. Deep Learning in Medical Image Analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, H.-P.; Samala, R.K.; Hadjiiski, L.M.; Zhou, C. Deep Learning in Medical Image Analysis. In Deep Learning in Medical Image Analysis; Lee, G., Fujita, H., Eds.; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Aziz, M.; Fatima, R.; Dong, C.; Lee-Smith, W.; Nawras, A. The impact of deep convolutional neural network-based artificial intelligence on colonoscopy outcomes: A systematic review with meta-analysis. J. Gastroenterol. Hepatol. 2020, 1–8. [Google Scholar] [CrossRef]
- Sánchez-Peralta, L.F.; Bote-Curiel, L.; Picón, A.; Sánchez-Margallo, F.M.; Pagador, J.B. Deep learning to find colorectal polyps in colonoscopy: A systematic literature review. Artif. Intell. Med. 2020, 108. [Google Scholar] [CrossRef] [PubMed]
- Nogueira-Rodríguez, A.; Domínguez-Carbajales, R.; López-Fernández, H.; Iglesias, Á.; Cubiella, J.; Fdez-Riverola, F.; Reboiro-Jato, M.; Glez-Peña, D. Deep Neural Networks approaches for detecting and classifying colorectal polyps. Neurocomputing 2020. [Google Scholar] [CrossRef]
- Min, J.K.; Kwak, M.S.; Cha, J.M. Overview of deep learning in gastrointestinal endoscopy. Gut Liver 2019, 13, 388–393. [Google Scholar] [CrossRef] [Green Version]
- Borgli, H.; Thambawita, V.; Smedsrud, P.; Hicks, S.; Jha, D.; Eskeland, S.; Randel, K.R.; Pogorelov, K.; Lux, M.; Dang-Nguyen, D.-T.; et al. HyperKvasir: A Comprehensive Multi-Class Image and Video Dataset for Gastrointestinal Endoscopy. Sci. Data 2020, 7. [Google Scholar] [CrossRef]
- Bernal, J.; Histace, A.; Masana, M.; Angermann, Q.; Sánchez-Montes, C.; de Miguel, C.R.; Hammami, M.; García-Rodríguez, A.; Córdova, H.; Romain, O. GTCreator: A flexible annotation tool for image-based datasets. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 191–201. [Google Scholar] [CrossRef]
- Tajbakhsh, N.; Jeyaseelan, L.; Li, Q.; Chiang, J.N.; Wu, Z.; Ding, X. Embracing Imperfect Datasets: A Review of Deep Learning Solutions for Medical Image Segmentation. Med. Image Anal. 2020, 63, 101693. [Google Scholar] [CrossRef] [Green Version]
- Vázquez, D.; Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; López, A.M.; Romero, A.; Drozdzal, M.; Courville, A. A Benchmark for Endoluminal Scene Segmentation of Colonoscopy Images. J. Healthc. Eng. 2017. [Google Scholar] [CrossRef]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Halvorsen, P.; de Lange, T.; Johansen, D.; Johansen, H.D. Kvasir-SEG: A Segmented Polyp Dataset. In Proceedings of the International Conference on Multimedia Modeling (MMM); Springer: Cham, Switzerland, 2020. [Google Scholar]
- Angermann, Q.; Bernal, J.; Sánchez-Montes, C.; Hammami, M.; Fernández-Esparrach, G.; Dray, X.; Romain, O.; Sánchez, F.J.; Histace, A. Towards Real-Time Polyp Detection in Colonoscopy Videos: Adapting Still Frame-Based Methodologies for Video Sequences Analysis. In Computer Assisted and Robotic Endoscopy and Clinical Image-Based Procedures; Springer: Cham, Switzerland, 2017; pp. 29–41. ISBN 978-3-319-67542-8. [Google Scholar]
- Participants in the Paris Workshop The Paris endoscopic classification of superficial neoplastic lesions: Esophagus, stomach, and colon. Gastrointest. Endosc. 2003, 58, S3–S43. [CrossRef]
- Endoscopic Classification Review Group Update on the Paris Classification of Superficial Neoplastic Lesions in the Digestive Tract. Endoscopy 2005, 37, 570–578. [CrossRef] [PubMed]
- Mesejo, P.; Pizarro, D.; Abergel, A.; Rouquette, O.; Beorchia, S.; Poincloux, L.; Bartoli, A. Computer-Aided Classification of Gastrointestinal Lesions in Regular Colonoscopy. IEEE Trans. Med. Imaging 2016, 35, 2051–2063. [Google Scholar] [CrossRef] [PubMed]
- Hattori, S.; Iwatate, M.; Sano, W.; Hasuike, N.; Kosaka, H.; Ikumoto, T.; Kotaka, M.; Ichiyanagi, A.; Ebisutani, C.; Hisano, Y.; et al. Narrow-band imaging observation of colorectal lesions using NICE classification to avoid discarding significant lesions. World J. Gastrointest. Endosc. 2014, 6, 600. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. arXiv 2018. [Google Scholar] [CrossRef] [Green Version]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention−MICCAI 2015. Lecture Notes in Computer Science; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; Volume 9351, pp. 234–241. ISBN 9783319245737. [Google Scholar]
- Chaurasia, A.; Culurciello, E. LinkNet: Exploiting encoder representations for efficient semantic segmentation. 2017 IEEE Vis. Commun. Image Process. VCIP 2018, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Peralta, L.F.; Picón, A.; Antequera-Barroso, J.A.; Ortega-Morán, J.F.; Sánchez-Margallo, F.M.; Pagador, J.B. Eigenloss: Combined PCA-Based Loss Function for Polyp Segmentation. Mathematics 2020, 8, 1316. [Google Scholar] [CrossRef]
- Yakubovskiy, P. Segmentation Models. Available online: https://github.com/qubvel/segmentation_models (accessed on 24 November 2020).
- Chollet, F. Keras. Available online: https://github.com/keras-team/keras (accessed on 24 November 2020).
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. CoRR 2015. [Google Scholar] [CrossRef] [Green Version]
- Abaza, A.; Harrison, M.A.; Bourlai, T. Quality metrics for practical face recognition. In Proceedings of the Proceedings of the 21st International Conference on Pattern Recognition (ICPR2012), Tsukuba, Japan, 11–15 November 2012; pp. 3103–3107. [Google Scholar]
- Tripathi, A.K.; Mukhopadhyay, S.; Dhara, A.K. Performance metrics for image contrast. In Proceedings of the 2011 International Conference on Image Information Processing, Shimla, India, 3–5 November 2011; pp. 1–4. [Google Scholar]
- Thambawita, V.; Jha, D.; Hammer, H.L.; Johansen, H.D.; Johansen, D.; Halvorsen, P.; Riegler, M.A. An Extensive Study on Cross-Dataset Bias and Evaluation Metrics Interpretation for Machine Learning Applied to Gastrointestinal Tract Abnormality Classification. ACM Trans. Comput. Healthc. 2020, 1, 1–29. [Google Scholar] [CrossRef]
- Lee, J.Y.; Jeong, J.; Song, E.M.; Ha, C.; Lee, H.J.; Koo, J.E.; Yang, D.H.; Kim, N.; Byeon, J.S. Real-time detection of colon polyps during colonoscopy using deep learning: Systematic validation with four independent datasets. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Sánchez-Peralta, L.F.; Sánchez-Margallo, F.M.; Bote Chacón, J.; Soria Gálvez, F.; López-Saratxaga, C.; Picón Ruiz, A.; Pagador, J.B. Is it necessary to improve the colorectal polyps databases for detection CAD systems based on deep learning? Br. J. Surg. 2018, 105, 5–14. [Google Scholar]
- Joskowicz, L.; Cohen, D.; Caplan, N.; Sosna, J. Inter-observer variability of manual contour delineation of structures in CT. Eur. Radiol. 2019, 29, 1391–1399. [Google Scholar] [CrossRef] [PubMed]
- Chlebus, G.; Meine, H.; Thoduka, S.; Abolmaali, N.; Van Ginneken, B.; Hahn, H.K.; Schenk, A. Reducing inter-observer variability and interaction time of MR liver volumetry by combining automatic CNN-based liver segmentation and manual corrections. PLoS ONE 2019, 14, e0217228. [Google Scholar] [CrossRef] [PubMed]
- Wickstrøm, K.; Kampffmeyer, M.; Jenssen, R. Uncertainty and Interpretability in Convolutional Neural Networks for Semantic Segmentation of Colorectal Polyps. Med. Image Anal. 2020, 60. [Google Scholar] [CrossRef] [PubMed]
- Nagendran, M.; Chen, Y.; Lovejoy, C.A.; Gordon, A.C.; Komorowski, M.; Harvey, H.; Topol, E.J.; Ioannidis, J.P.A.; Collins, G.S.; Maruthappu, M. Artificial intelligence versus clinicians: Systematic review of design, reporting standards, and claims of deep learning studies in medical imaging. BMJ 2020, 368, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Category | Items | # Lesions | # WL Frames | # NBI Frames |
|---|---|---|---|---|
| Paris Classification | Protruded lesions: 0-Ip | 10 | 191 | 193 |
| Protruded lesions: 0-Ips | 7 | 193 | 107 | |
| Protruded lesions: 0-Is | 16 | 387 | 253 | |
| Flat elevated lesions: 0-IIa | 29 | 928 | 436 | |
| Flat elevated lesions:0-IIa/c | 4 | 83 | 91 | |
| Flat lesions: 0-IIb | 2 | 85 | 21 | |
| N/A | 8 | 264 | 201 | |
| NICE classification | Type 1 | 17 | 404 | 284 |
| Type 2 | 50 | 1454 | 782 | |
| Type 3 | 8 | 264 | 201 | |
| N/A | 1 | 9 | 35 | |
| Diagnosis | Adenocarcinoma | 8 | 264 | 201 |
| Adenoma | 50 | 1454 | 782 | |
| Hyperplasia | 17 | 404 | 284 | |
| N/A | 1 | 9 | 35 | |
| Histological stratification | High grade dysplasia | 12 | 409 | 168 |
| Hyperplasia | 13 | 312 | 225 | |
| Invasive adenocarcinoma | 8 | 264 | 201 | |
| Low grade dysplasia | 1 | 13 | 16 | |
| No dysplasia | 41 | 1124 | 657 | |
| N/A | 1 | 9 | 35 |
| Category | Items | Train Set | Validation Set | Test Set |
|---|---|---|---|---|
| Image type | WL | 1382 | 558 | 192 |
| NBI | 821 | 340 | 141 | |
| Paris Classification | Protruded lesions: 0-Ip | 274 | 81 | 29 |
| Protruded lesions: 0-Ips | 245 | 41 | 14 | |
| Protruded lesions: 0-Is | 433 | 176 | 31 | |
| Flat elevated lesions: 0-IIa | 1052 | 263 | 49 | |
| Flat elevated lesions:0-IIa/c | 27 | 122 | 25 | |
| Flat lesions: 0-IIb | - | 48 | 58 | |
| N/A | 172 | 166 | 127 | |
| NICE classification | Type 1 | 435 | 139 | 114 |
| Type 2 | 1552 | 592 | 92 | |
| Type 3 | 172 | 166 | 127 | |
| N/A | 44 | - | - | |
| Diagnosis | Adenocarcinoma | 172 | 166 | 127 |
| Adenoma | 1552 | 592 | 92 | |
| Hyperplasia | 435 | 139 | 114 | |
| N/A | 44 | - | - | |
| Histological stratification | High grade dysplasia | 360 | 217 | - |
| Hyperplasia | 342 | 139 | 56 | |
| Invasive adenocarcinoma | 172 | 166 | 127 | |
| Low grade dysplasia | - | - | 29 | |
| No dysplasia | 1285 | 375 | 121 | |
| N/A | 44 | - | - |
| CVC-EndoSceneStill | Kvasir-SEG | PICCOLO | |
|---|---|---|---|
| Void area (%) | 23.73 ± 5.57 (27.83 − 14.62) | 15.23 ± 4.82 (28.44 − 6.16) | 34.14 ± 0.33 (34.33 − 33.20) |
| Polyp area relative to the valid area (%) | 12.50 ± 11.49 (66.15 − 0.75) | 17.36 ± 15.65 (83.66 − 0.61) | 20.45 ± 20.68 (86.11 − 0.00) |
| Mean value of brightness channel in HSV | 0.560 ± 0.006 (1.000 − 0.000) | 0.622 ± 0.003 (1.000 − 0.000) | 0.532 ± 0.078 (1.000 − 0.000) |
| Histogram flatness measure | 0.858 ± 0.121 (0.959 − 0.000) | 0.419 ± 0.443 (0.962 − 0.000) | 0.855 ± 0.215 (0.970 − 0.000) |
| Histogram spread | 0.252 ± 0.088 (0.520 − 0.076) | 0.218 ± 0.070 (0.432 − 0.075) | 0.214 ± 0.086 (0.452 − 0.066) |
| Dataset for Training/Validation Set | Network | Dataset for Test Set | |||
|---|---|---|---|---|---|
| CVC-EndoSceneStill | Kvasir-SEG | PICCOLO | All | ||
| CVC-EndoSceneStill | U-Net + VGG16 | 56.80 ± 36.09 | 46.29 ± 29.36 | 32.82 ± 34.81 | 42.69 ± 35.13 |
| U-Net + Densenet121 | 67.31 ± 34.47 | 56.12 ± 34.29 | 38.76 ± 39.05 | 50.88 ± 38.51 | |
| LinkNet + VGG16 | 62.77 ± 33.81 | 51.13 ± 27.67 | 30.55 ± 33.57 | 44.51 ± 34.86 | |
| LinkNet + Densenet121 | 72.16 ± 30.93 | 56.69 ± 33.68 | 39.52 ± 37.90 | 52.63 ± 37.53 | |
| Kvasir-SEG | U-Net + VGG16 | 32.44 ± 38.79 | 64.23 ± 29.78 | 28.51 ± 35.12 | 39.50 ± 37.98 |
| U-Net + Densenet121 | 71.82 ± 29.87 | 74.13 ± 23.40 | 44.78 ± 38.73 | 59.87 ± 35.72 | |
| LinkNet + VGG16 | 58.62 ± 36.18 | 72.53 ± 23.92 | 40.43 ± 36.41 | 54.04 ± 35.99 | |
| LinkNet + Densenet121 | 69.10 ± 32.53 | 74.52 ± 22.81 | 44.92 ± 37.37 | 59.35 ± 35.33 | |
| PICCOLO | U-Net + VGG16 | 47.76 ± 36.46 | 52.64 ± 30.41 | 58.74 ± 36.06 | 54.24 ± 34.93 |
| U-Net + Densenet121 | 62.41 ± 34.78 | 65.33 ± 30.66 | 64.01 ± 36.23 | 63.97 ± 34.35 | |
| LinkNet + VGG16 | 54.58 ± 35.63 | 58.99 ± 30.40 | 54.46 ± 38.88 | 55.76 ± 35.87 | |
| LinkNet + Densenet121 | 64.18 ± 33.04 | 59.61 ± 33.80 | 60.14 ± 38.31 | 61.02 ± 35.79 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Peralta, L.F.; Pagador, J.B.; Picón, A.; Calderón, Á.J.; Polo, F.; Andraka, N.; Bilbao, R.; Glover, B.; Saratxaga, C.L.; Sánchez-Margallo, F.M. PICCOLO White-Light and Narrow-Band Imaging Colonoscopic Dataset: A Performance Comparative of Models and Datasets. Appl. Sci. 2020, 10, 8501. https://doi.org/10.3390/app10238501
Sánchez-Peralta LF, Pagador JB, Picón A, Calderón ÁJ, Polo F, Andraka N, Bilbao R, Glover B, Saratxaga CL, Sánchez-Margallo FM. PICCOLO White-Light and Narrow-Band Imaging Colonoscopic Dataset: A Performance Comparative of Models and Datasets. Applied Sciences. 2020; 10(23):8501. https://doi.org/10.3390/app10238501
Chicago/Turabian StyleSánchez-Peralta, Luisa F., J. Blas Pagador, Artzai Picón, Ángel José Calderón, Francisco Polo, Nagore Andraka, Roberto Bilbao, Ben Glover, Cristina L. Saratxaga, and Francisco M. Sánchez-Margallo. 2020. "PICCOLO White-Light and Narrow-Band Imaging Colonoscopic Dataset: A Performance Comparative of Models and Datasets" Applied Sciences 10, no. 23: 8501. https://doi.org/10.3390/app10238501
APA StyleSánchez-Peralta, L. F., Pagador, J. B., Picón, A., Calderón, Á. J., Polo, F., Andraka, N., Bilbao, R., Glover, B., Saratxaga, C. L., & Sánchez-Margallo, F. M. (2020). PICCOLO White-Light and Narrow-Band Imaging Colonoscopic Dataset: A Performance Comparative of Models and Datasets. Applied Sciences, 10(23), 8501. https://doi.org/10.3390/app10238501