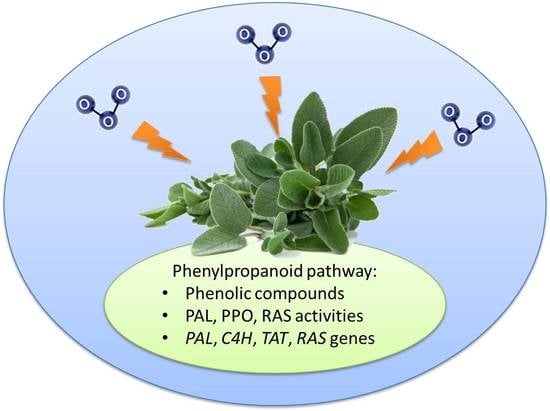
The Biosynthesis of Phenolic Compounds Is an Integrated Defence Mechanism to Prevent Ozone Injury in Salvia officinalis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Biochemical Analyses
2.2.1. Metabolites Involved in the Phenylpropanoid Pathway
2.2.2. Enzymes Involved in Phenolic Metabolism
2.3. Molecular Analyses
2.3.1. qRT-PCR Primer Design
2.3.2. RNA Extraction and Relative Expression Analyses
2.4. Statistical Analyses
3. Results
3.1. Metabolites Involved in the Phenylpropanoid Pathway
3.2. Enzymes Involved in the Phenylpropanoid Pathway
3.3. Gene Expression Analyses
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Soares, C.; Carvalho, M.E.; Azevedo, R.A.; Fidalgo, F. Plants facing oxidative challenges—A little help from the antioxidant networks. Environ. Exp. Bot. 2019, 161, 4–25. [Google Scholar] [CrossRef]
- Huang, H.; Ullah, F.; Zhou, D.-X.; Yi, M.; Zhao, Y. Mechanisms of ROS regulation of plant development and stress responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef] [PubMed]
- Isah, T. Stress and defense responses in plant secondary metabolites production. Biol. Res. 2019, 52, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef] [Green Version]
- Cheynier, V.; Comte, G.; Davies, K.M.; Lattanzio, V.; Martens, S. Plant phenolics: Recent advances on their biosynthesis, genetics, and ecophysiology. Plant Physiol. Biochem. 2013, 72, 1–20. [Google Scholar] [CrossRef]
- Rice-Evans, C.A.; Miller, N.J.; Paganga, G. Structure-antioxidant activity relationships of flavonoids and phenolic acids. Free Radic. Biol. Med. 1996, 20, 933–956. [Google Scholar] [CrossRef]
- Olszowy, M. What is responsible for antioxidant properties of polyphenolic compounds from plants? Plant Physiol. Biochem. 2019, 14, 135–143. [Google Scholar] [CrossRef]
- Döring, A.S.; Pellegrini, E.; Della Bartola, M.; Nali, C.; Lorenzini, G.; Petersen, M. How do background ozone concentrations affect the biosynthesis of rosmarinic acid in Melissa officinalis? J. Plant Physiol. 2014, 171, 35–41. [Google Scholar] [CrossRef]
- Vergine, M.; Nicolì, F.; Negro, C.; Luvisi, A.; Nutricati, E.; Accogli, R.A.; Sabella, E.; Miceli, A. Phytochemical profiles and antioxidant activity of Salvia species from Southern Italy. Rec. Nat. Prod. 2019, 13, 205–215. [Google Scholar] [CrossRef]
- Lu, Y.; Foo, L.Y. Polyphenolics of Salvia—A review. Phytochemistry 2002, 59, 117–140. [Google Scholar] [CrossRef]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Pandey, A.K. Chemistry and biological activities of flavonoids: An overview. Sci. World J. 2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorenzini, G.; Nali, C. Ozone and plant life: The Italian state-of-the-art. Environ. Sci. Pollut. Res. 2018, 25, 8069–8073. [Google Scholar] [CrossRef] [Green Version]
- European Environment Agency (EEA). Air Quality in Europe. EEA Report 10/2019. Available online: https://www.eea.europa.eu/publications/air-quality-in-europe-2019 (accessed on 21 October 2019).
- Cotrozzi, L.; Pellegrini, E.; Nali, C.; Lorenzini, G. Climate change, ozone and plant life. Agrochimica 2019, 181–188. [Google Scholar]
- Pellegrini, E.; Francini, A.; Nali, C.; Lorenzini, G. Ecophysiological and antioxidant traits of Salvia officinalis under ozone stress. Environ. Sci. Pollut. Res. 2015, 22, 13083–13093. [Google Scholar] [CrossRef]
- Emberson, L.D.; Pleijel, H.; Ainsworth, E.A.; van den Berg, M.; Ren, W.; Osborne, S.; Mills, G.; Pandey, D.; Dentener, F.; Büker, P.; et al. Ozone effects on crops and consideration in crop models. Eur. J. Agron. 2018, 100, 19–34. [Google Scholar] [CrossRef]
- Marchica, A.; Lorè, S.; Cotrozzi, L.; Lorenzini, G.; Nali, C.; Pellegrini, E.; Remorini, D. Early detection of sage (Salvia officinalis L.) responses to ozone using reflectance spectroscopy. Plants 2019, 8, 346. [Google Scholar] [CrossRef] [Green Version]
- Fini, A.; Guidi, L.; Ferrini, F.; Brunetti, C.; Di Ferdinando, M.; Biricolti, S.; Pollastri, S.; Calamai, L.; Tattini, M. Drought stress has contrasting effects on antioxidant enzymes activity and phenylpropanoid biosynthesis in Fraxinus ornus leaves: An excess light stress affair? J. Plant Physiol. 2012, 169, 929–939. [Google Scholar] [CrossRef]
- Ragusa, A.; Centonze, C.; Grasso, M.E.; Latronico, M.F.; Mastrangelo, P.F.; Sparascio, F.; Maffia, M. HPLC analysis of phenols in Negroamaro and Primitivo red wines from Salento. Foods 2019, 8, 45. [Google Scholar] [CrossRef] [Green Version]
- Leuschner, M. Untersuchungen zu Phenoloxidasen aus Zellkulturen von Lycopersicum esculentum Mill. Doctoral Thesis, Philipps-Universität Marburg, Marburg, Germany, 2002. [Google Scholar]
- Berger, A.; Meinhard, J.; Petersen, M. Rosmarinic acid synthase is a new member of the superfamily of BAHD acyltransferases. Planta 2006, 224, 1503–1510. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 2002–20007. [Google Scholar] [CrossRef]
- Sgarbi, E.; Medeghini Bonatti, P.; Baroni Fornasiero, R.; Lins, A. Differential sensitivity to ozone in two selected cell lines from grape leaf. J. Plant Physiol. 1999, 154, 119–126. [Google Scholar] [CrossRef]
- Pasqualini, S.; Piccioni, C.; Reale, L.; Ederli, L.; Della Torre, G.; Ferranti, F. Ozone-induced cell death in tobacco cultivar Bel W3 plants. The role of programmed cell death in lesion formation. Plant Physiol. 2003, 133, 1122–1134. [Google Scholar] [CrossRef] [Green Version]
- Pellegrini, E.; Hoshika, Y.; Dusart, N.; Cotrozzi, L.; Gérard, J.; Nali, C.; Vaultier, M.N.; Jolivet, Y.; Lorenzini, G.; Paoletti, E. Antioxidative responses of three oak species under ozone and water stress conditions. Sci. Total Environ. 2019, 647, 390–399. [Google Scholar] [CrossRef] [PubMed]
- Larson, R.A. The antioxidants of higher plants. Phytochemistry 1988, 27, 969–978. [Google Scholar] [CrossRef]
- Widhalm, J.R.; Dudareva, N. A familiar ring to it: Biosynthesis of plant benzoic acids. Mol. Plant 2015, 8, 83–97. [Google Scholar] [CrossRef] [Green Version]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Agati, G.; Tattini, M. Multiple functional roles of flavonoids in photoprotection. New Phytol. 2010, 186, 786–793. [Google Scholar] [CrossRef]
- Mosadegh, H.; Trivellini, A.; Ferrante, A.; Lucchesini, M.; Vernieri, P.; Mensuali, A. Applications of UV-B lighting to enhance phenolic accumulation of sweet basil. Sci. Hortic. 2018, 229, 107–116. [Google Scholar] [CrossRef]
- Yoshikawa, M.; Luo, W.; Tanaka, G.; Konishi, Y.; Matsuura, H.; Takahashi, K. Wounding stress induces phenylalanine ammonia lyase, leading to the accumulation of phenylpropanoids in the model liveworth Marchantia polymorpha. Phytochemistry 2018, 155, 30–36. [Google Scholar] [CrossRef]
- Grulke, N.E.; Paoletti, E.; Heat, R.L. Chronic vs. short-term acute O3 exposure effects on nocturnal transpiration in two Californian Oaks. Sci. World J. 2007, 7, 134–140. [Google Scholar] [CrossRef] [Green Version]
- Cotrozzi, L. The effects of tropospheric ozone on oaks: A global meta analysis. Sci. Total Environ. 2020, 756, 143795. [Google Scholar] [CrossRef]
- Wellburn, F.A.M.; Wellburn, A.R. Variable patterns of antioxidant protection but similar ethene emission differences in several ozone-sensitive and ozone-tolerant plant selections. Plant Cell Environ. 1996, 19, 754–760. [Google Scholar] [CrossRef]
- Agati, G.; Brunetti, C.; Fini, A.; Gori, A.; Guidi, L.; Landi, M.; Sebastiani, F.; Tattini, M. Are flavonoids effective antioxidants in plants? Twenty years of our investigation. Antioxidant 2020, 9, 1098. [Google Scholar] [CrossRef]
- Ahuja, L.; Kissen, R.; Bones, A.M. Phytoalexins in defence against pathogens. Trends Plant Sci. 2012, 17, 73–90. [Google Scholar] [CrossRef]
- Kangasjarvi, J.; Jaspers, P.; Kollist, H. Signalling and cell death in ozone-exposed plants. Plant Cell Environ. 2005, 28, 1021–1035. [Google Scholar] [CrossRef]
- Hahlbrock, K.; Scheel, D. Physiology and molecular biology of phenylpropanoid metabolism. Annu. Rev. Plant Biol. 1989, 40, 347–369. [Google Scholar] [CrossRef]
- Taranto, F.; Pasqualone, A.; Mangini, G.; Tripodi, P.; Miazzi, M.M.; Pavan, S.; Montemurro, C. Polyphenol oxidases in crops: Biochemical, physiological and genetic aspects. Int. J. Mol. Sci. 2017, 18, 337. [Google Scholar] [CrossRef]
- Petersen, M.; Alfermann, A.W. Two new enzymes of rosmarinic acid biosynthesis from cell cultures of Coleus blumei: Hydroxyphenylpyruvate reductase and rosmarinic acid synthase. Z. Nat. C 1988, 43, 501–504. [Google Scholar] [CrossRef]
- Rao, M.V.; Hale, B.A.; Ormrod, D.P. Amelioration of ozone-induced oxidative damage in wheat plants grown under high carbon dioxide. Plant Physiol. 1995, 109, 421–432. [Google Scholar] [CrossRef]
- Zhao, D.L.C.; Verpoorte, R. Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol. Adv. 2005, 23, 283–333. [Google Scholar] [CrossRef]
- Wang, B.; Sun, W.; Li, Q.; Li, Y.; Luo, H.; Song, J.; Sun, C.; Qian, J.; Zhu, Y.; Hayward, A.; et al. Genome-wide identification of phenolic acid biosynthetic genes in Salvia miltiorrhiza. Planta 2015, 241, 711–725. [Google Scholar] [CrossRef]
- Ejtahed, R.; Radjabian, T.; Hoseini Tafreshi, S.A. Expression analysis of phenylalanine ammonia lyase gene and rosmarinic acid production in Salvia officinalis and Salvia virgata shoots under salicylic acid elicitation. Appl. Biochem. Biotechnol. 2015, 176, 1846–1858. [Google Scholar] [CrossRef]
- Raes, J.; Rohde, A.; Christensen, J.H.; Van de Peer, Y.; Boerjan, W. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 2003, 133, 1051–1071. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Wang, H.; Cheng, X.; Su, X.; Zhao, Y.; Jaing, T.; Jin, Q.; Lin, Y.; Cai, Y. Comparative genomic analysis of the PAL genes in five Rosaceae species and functional identification of Chinese white pear. Peer J. 2019, 7, e8064. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.Y.; Yu, H.N.; Gao, S.; Wu, Y.F.; Cheng, A.X.; Lou, H.X. The isolation and functional characterization of three liverwort genes encoding cinnamate 4-hydroxylase. Plant Physiol. Biochem. 2014, 117, 42–50. [Google Scholar] [CrossRef]
- Gottardini, E.; Cristofori, A.; Pellegrini, E.; La Porta, N.; Nali, C.; Baldi, P.; Sablok, G. Suppression subtractive hybridization and NGS reveal differential transcriptome expression profiles in wayfaring tree (Viburnum lantana L.) treated with ozone. Front. Plant Sci. 2016, 7, 713. [Google Scholar] [CrossRef] [Green Version]
- Song, Z.; Li, X. Expression profiles of rosmarinic acid biosynthesis genes in two Salvia miltiorrhiza lines with differing water-soluble phenolic contents. Ind. Crops Prod. 2015, 71, 24–30. [Google Scholar] [CrossRef]
- Kim, K.H.; Janiak, V.; Petersen, M. Purification, cloning and functional expression of hydroxyphenylpyruvate reductase involved in rosmarinic acid biosynthesis in cell cultures of Coleus blumei. Plant Mol. Biol. 2004, 54, 311–323. [Google Scholar] [CrossRef]
- Wang, M.; Toda, K.; Block, A.; Maeda, H.A. TAT1 and TAT2 tyrosine aminotransferases have both distinct and shared functions in tyrosine metabolism and degradation in Arabidopsis thaliana. J. Biol. Chem. 2019, 294, 3563–3576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trócsányi, E.; György, Z.; Zámboriné-Németh, É. New insights into rosmarinic acid biosynthesis based on molecular studies. Curr. Plant Biol. 2020, 23, 100162. [Google Scholar] [CrossRef]





| Primers | Sequences |
|---|---|
| EF1α | F: 5′-ACAACCCTGAGAAGATCCC-3′ |
| R: 5′-GCACAGTTCCAATACCACCAAT-3′ | |
| Actin | F: 5′-TCTCTTGACAGAAGCCCCTCT-3′ |
| R: 5′-GATGGGGACTGTATGGCTGA-3′ | |
| PAL | F: 5′-GAAGAACACCGTGAGCCAAG-3′ |
| R:5′-GCTCACACTCTTCTCCCCTT-3′ | |
| C4H | F: 5′-ATCTGAACCACCGCAACCTC-3′ |
| R: 5′-GTGAACACCATGTCCTGACC-3′ | |
| TAT | F: 5′-GCACCTAAAGAAGATTGCTGAG-3′ |
| R: 5′-ATTGATGACCAACCAACCAAGG-3′ | |
| RAS | F:5′-ACTACTTGAGGTCGTCGCTC-3′ |
| R:5′-CGGATTTGGCAGGAGATAGC-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marchica, A.; Cotrozzi, L.; Detti, R.; Lorenzini, G.; Pellegrini, E.; Petersen, M.; Nali, C. The Biosynthesis of Phenolic Compounds Is an Integrated Defence Mechanism to Prevent Ozone Injury in Salvia officinalis. Antioxidants 2020, 9, 1274. https://doi.org/10.3390/antiox9121274
Marchica A, Cotrozzi L, Detti R, Lorenzini G, Pellegrini E, Petersen M, Nali C. The Biosynthesis of Phenolic Compounds Is an Integrated Defence Mechanism to Prevent Ozone Injury in Salvia officinalis. Antioxidants. 2020; 9(12):1274. https://doi.org/10.3390/antiox9121274
Chicago/Turabian StyleMarchica, Alessandra, Lorenzo Cotrozzi, Rebecca Detti, Giacomo Lorenzini, Elisa Pellegrini, Maike Petersen, and Cristina Nali. 2020. "The Biosynthesis of Phenolic Compounds Is an Integrated Defence Mechanism to Prevent Ozone Injury in Salvia officinalis" Antioxidants 9, no. 12: 1274. https://doi.org/10.3390/antiox9121274
APA StyleMarchica, A., Cotrozzi, L., Detti, R., Lorenzini, G., Pellegrini, E., Petersen, M., & Nali, C. (2020). The Biosynthesis of Phenolic Compounds Is an Integrated Defence Mechanism to Prevent Ozone Injury in Salvia officinalis. Antioxidants, 9(12), 1274. https://doi.org/10.3390/antiox9121274