Hybrid Heme Peroxidases from Rice Blast Fungus Magnaporthe oryzae Involved in Defence against Oxidative Stress
Abstract
:1. Introduction
2. Materials and Methods
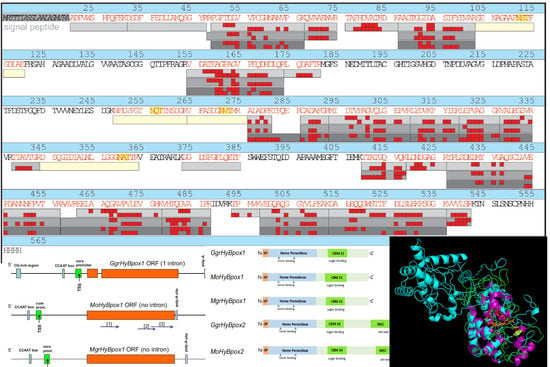
2.1. Bioinformatic and Phylogenetic Analysis of Hybrid Peroxidases
2.2. Cultivation of the Fungus and Detection of Native Expression of MohyBpox1 and MohyBpox2 Gene Paralogs
2.3. Heterologous Expression of MoHyBPOX1 in Pichia pastoris BG11 (AOX1) Strain
2.4. Purification of Recombinantly Produced MoHyBPOX1
2.5. Deglycosylation of Purified Hybrid Peroxidase
2.6. Protein Identification and Peptide Analysis Using LC-ESI-MS
2.7. N-Glycan Release and LC-ESI-MS Analysis of Free N-Glycans
2.8. UV-Vis and Electronic Circular Dichroism Spectroscopy
2.9. Electron Paramagnetic Resonance
2.10. Thermodynamic and Kinetics of Cyanide Binding
2.11. Determination of the Peroxidatic Activity
2.12. Detection of Binding of Soluble Starch on Purified MoHyBPOX1 Protein
3. Results and Discussion
3.1. Phylogenomic Analysis and Quantification of Native Expression for MohyBpox1 and MohyBpox2 Gene Paralogs with RT-qPCR Methodology
3.2. Heterologous Expression of Recombinant MoHyBPOX1 in Pichia pastoris
3.3. Spectroscopic Properties of Hybrid B Peroxidase
3.4. Unique Catalytic Properties of Hybrid B Peroxidase
3.5. Binding of Soluble Sugars
3.6. Structural Peculiarities of Hybrid Peroxidases as Fusion Proteins
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Savelli, B.; Li, Q.; Webber, M.; Jemmat, A.M.; Robitaille, A.; Zamocky, M.; Mathé, C.; Dunand, C. RedoxiBase: A database for ROS homeostasis regulated proteins. Redox Boil. 2019, 26, 101247. [Google Scholar] [CrossRef] [PubMed]
- Zámocký, M.; Hofbauer, S.; Schaffner, I.; Gasselhuber, B.; Nicolussi, A.; Soudi, M.; Pirker, K.F.; Furtmüller, P.G.; Obinger, C. Independent evolution of four heme peroxidase superfamilies. Arch. Biochem. Biophys. 2015, 574, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Zamocky, M.; Janeček, Š.; Obinger, C. Fungal Hybrid B heme peroxidases—Unique fusions of a heme peroxidase domain with a carbohydrate-binding domain. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zamocky, M.; Gasselhuber, B.; Furtmüller, P.G.G.; Obinger, C. Turning points in the evolution of peroxidase–catalase superfamily: Molecular phylogeny of hybrid heme peroxidases. Cell. Mol. Life Sci. 2014, 71, 4681–4696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reynolds, K.A.; Russ, W.P.; Socolich, M.; Ranganathan, R. Evolution-Based Design of Proteins. Methods Enzymol. 2013, 523, 213–235. [Google Scholar] [CrossRef] [PubMed]
- Adak, S.; Datta, A.K. Leishmania major encodes an unusual peroxidase that is a close homologue of plant ascorbate peroxidase: A novel role of the transmembrane domain. Biochem. J. 2005, 390, 465–474. [Google Scholar] [CrossRef] [Green Version]
- Ishikawa, T.; Tajima, N.; Nishikawa, H.; Gao, Y.; Rapolu, M.; Shibata, H.; Sawa, Y.; Shigeoka, S. Euglena gracilis ascorbate peroxidase forms an intramolecular dimeric structure: Its unique molecular characterization. Biochem. J. 2010, 426, 125–134. [Google Scholar] [CrossRef] [Green Version]
- Krueger, T.; Fisher, P.L.; Becker, S.; Pontasch, S.; Dove, S.; Hoegh-Guldberg, O.; Leggat, W.; Davy, S.K. Transcriptomic characterization of the enzymatic antioxidants FeSOD, MnSOD, APX and KatG in the dinoflagellate genus Symbiodinium. BMC Evol. Boil. 2015, 15, 48. [Google Scholar] [CrossRef] [Green Version]
- Zamocky, M.; Tafer, H.; Chovanová, K.; Lopandic’, K.; Kamlárová, A.; Obinger, C. Genome sequence of the filamentous soil fungus Chaetomium cochliodes reveals abundance of genes for heme enzymes from all peroxidase and catalase superfamilies. BMC Genom. 2016, 17, 763. [Google Scholar] [CrossRef] [Green Version]
- Bellora, N.; Moliné, M.; David-Palma, M.; Coelho, M.A.; Hittinger, C.T.; Sampaio, J.-P.; Gonçalves, P.; Libkind, D. Comparative genomics provides new insights into the diversity, physiology, and sexuality of the only industrially exploited tremellomycete: Phaffia rhodozyma. BMC Genomics 2016, 17, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Smit, S.; Derks, M.F.L.; Bervoets, S.; Fahal, A.H.; Van Leeuwen, W.; Van Belkum, A.; Van De Sande, W.W.J. Genome Sequence of Madurella mycetomatis mm55, Isolated from a Human Mycetoma Case in Sudan. Genome Announc. 2016, 4, 00418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, K.; Saunders, D.G.O.; Mitsuoka, C.; Natsume, S.; Kosugi, S.; Saitoh, H.; Inoue, Y.; Chuma, I.; Tosa, Y.; Cano, L.M.; et al. Host specialization of the blast fungus Magnaporthe oryzae is associated with dynamic gain and loss of genes linked to transposable elements. BMC Genomics 2016, 17, 370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callaway, E. Devastating wheat fungus appears in Asia for first time. Natural 2016, 532, 421–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doehlemann, G.; Ökmen, B.; Zhu, W.; Sharon, A. Plant Pathogenic Fungi. Microbiol. Spectr. 2017, 5, FUNK-0023. [Google Scholar] [CrossRef]
- Döhlemann, G.; Hemetsberger, C. Apoplastic immunity and its suppression by filamentous plant pathogens. New Phytol. 2013, 198, 1001–1016. [Google Scholar] [CrossRef]
- Solovyev, V.V.; Kosarev, P.; Seledsov, I.; Vorobyev, D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006, 7, S10. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence aligment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Pierleoni, A.; Martelli, P.L.; Fariselli, P.; Casadio, R. BaCelLo: A balanced subcellular localization predictor. Bioinformatics 2006, 22, e408–e416. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Boil. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.E.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Boil. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, N.; Hensten-Pettersen, A. Salivary Amylase. Caries Res. 1970, 4, 193–199. [Google Scholar] [CrossRef]
- Oide, S.; Tanaka, Y.; Watanabe, A.; Inui, M. Carbohydrate-binding property of a cell wall integrity and stress response component (WSC) domain of an alcohol oxidase from the rice blast pathogen Pyricularia oryzae. Enzym. Microb. Technol. 2019, 125, 13–20. [Google Scholar] [CrossRef]
- Garcia-Arellano, H. Chapter 13—A Compendium of Biophysical-Chemical Properties of Peroxidases. In Biocatalysis Based on Heme Peroxidases; Torres, E., Ayala, M., Eds.; Springer: Berlin, Germany, 2010; pp. 335–352. [Google Scholar]
- Zamocky, M.; Furtmüller, P.G.G.; Bellei, M.; Battistuzzi, G.; Stadlmann, J.; Vlasits, J.; Obinger, C. Intracellular catalase/peroxidase from the phytopathogenic rice blast fungus Magnaporthe grisea: Expression analysis and biochemical characterization of the recombinant protein. Biochem. J. 2009, 418, 443–451. [Google Scholar] [CrossRef] [Green Version]
- Zamocky, M.; Droghetti, E.; Bellei, M.; Gasselhuber, B.; Pabst, M.; Furtmüller, P.G.; Battistuzzi, G.; Smulevich, G.; Obinger, C. Eukaryotic extracellular catalase–peroxidase from Magnaporthe grisea—Biophysical/chemical characterization of the first representative from a novel phytopathogenic KatG group. Biochimie 2012, 94, 673–683. [Google Scholar] [CrossRef] [Green Version]
- Plieth, C.; Vollbehr, S. Calcium promotes activity and confers heat stability on plant peroxidases. Plant Signal. Behav. 2012, 7, 650–660. [Google Scholar] [CrossRef] [Green Version]
- Rekik, H.; Jaouadi, N.Z.; Bouacem, K.; Zenati, B.; Kourdali, S.; Badis, A.; Annane, R.; Bouanane-Darenfed, A.; Bejar, S.; Jaouadi, B. Physical and enzymatic properties of a new manganese peroxidase from the white-rot fungus Trametes pubescens strain i8 for lignin biodegradation and textile-dyes biodecolorization. Int. J. Boil. Macromol. 2019, 125, 514–525. [Google Scholar] [CrossRef]
- Krainer, F.W.; Pletzenauer, R.; Rossetti, L.; Herwig, C.; Glieder, A.; Spadiut, O. Purification and basic biochemical characterization of 19 recombinant plant peroxidase isoenzymes produced in Pichia pastoris. Protein Expr. Purif. 2014, 95, 104–112. [Google Scholar] [CrossRef] [Green Version]
- Koduri, R.S.; Tien, M. Oxidation of Guaiacol by Lignin Peroxidase. J. Boil. Chem. 1995, 270, 22254–22258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saez-Jimenez, V.; Fernández-Fueyo, E.; Medrano, F.J.; Romero, A.; Martínez, A.T.; Ruiz-Duenas, F.J. Improving the pH-stability of Versatile Peroxidase by Comparative Structural Analysis with a Naturally-Stable Manganese Peroxidase. PLoS ONE 2015, 10, e0140984. [Google Scholar] [CrossRef]
- Lauber, C.; Schwarz, T.; Nguyen, Q.K.; Lorenz, P.; Lochnit, G.; Zorn, H. Identification, heterologous expression and characterization of a dye-decolorizing peroxidase of Plerutorus sapidus. AMB Expr. 2017, 7, 164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Yan, R.; Roy, A.; Xu, N.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2014, 12, 7–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [Green Version]







| Gene Identification (According to RedoxiBase) | GenBank Accessions (ID #) |
|---|---|
| MohyBpox1 | CD037344, JZ969880; JZ969881; JZ969916; JZ969917; JZ969918; JZ969919; JZ969979; JZ970396; JZ970397; JZ970398; JZ970399; JZ970417; JZ970418 |
| MohyBpox2 | JZ970248; JZ970249; JZ970250; JZ970400; JZ970401; JZ970402; JZ970251 |
| Peroxidase Identification | α-Helix (%) | β-Strand (%) | Other (%) * |
|---|---|---|---|
| MoHyBPOX1—this work | 39.3 | 14.2 | 46.3 |
| PsAPX (PDB code: 1apx)—Family I representative | 47.4 | 1.6 | 51.0 |
| PcMnP (1mnp)—Family II representative | 31.1 | 3.4 | 65.5 |
| HRP (1atj)—Family III representative | 43.5 | 2.0 | 54.5 |
| Substrate | E0´ of Sub. (V) | Detection at WL (nm) | pH Optimum | Specific Activity (U/mg) |
|---|---|---|---|---|
| ABTS | 1.08 | 414 | 6.5 | 217.8 ± 8.3 |
| Guaiacol | 0.43 | 470 | 6.0 | 8.0 ± 1.1 |
| L-DOPA | 0.47 | 470 | 6.5 | 103.1 ± 6.6 |
| 5-Aminosalicylate | 0.54 | 470 | 6.5 | 188.4 ±10.3 |
| Catechol | 1.20 | 350 | 5.5 | 5.7 ± 0.8 |
| Resorcinol | 1.70 | 290 | 5.0 | 1.1 ± 0.4 |
| Pyrogallol | 0.56 | 430 | 5.5 | 40.2 ± 0.7 |
| 3,3,5,5-Tetramethylbenzidine | 0.60 | 652 | 4.5 | 21.1 ± 1.5 |
| Ascorbate | 0.28 | 290 | 6.0 | 7.2 ± 0.1 |
| Cytochrome c | 0.24 | 550 | 7.0 | 0 * |
| Mn2+ | 1.51 | 238 | 8.5 | 7.2 ± 0.3 |
| L-Tyrosine | 0.93 | fluorescence | 7.5 | 621.6 ± 39.2 FU ** |
| Electron Donor | KM (µM) | kcat (s−1) | kcat/KM (M−1 s−1) |
|---|---|---|---|
| ABTS | 11.2 ± 1.0 | 106.3 ± 7.4 | 9.52 × 106 |
| L-DOPA | 69.7 ± 2.8 | 24.5 ± 5.4 | 3.51 × 105 |
| Pyrogallol | 29.3 ± 1.3 | 1.1 ± 0.2 | 3.62 × 104 |
| Guaiacol | 97.3 ± 1.9 | 0.5 ± 0.1 | 5.16 × 103 |
| TMB | 69.2 ± 0.7 | 0.2 ± 0.02 | 2.75 × 103 |
| Ascorbate | 250.2 ± 2.3 | 1.2 ± 0.2 | 4.92 × 103 |
| Mn2+ | 762.4 ± 6.9 | 4.5 ± 0.3 | 5.96 × 103 |
| Protein | Starch Binding (µM/µMprot) * |
|---|---|
| MoHyBPOX1 | 324.15 ± 38.04 |
| Cytochrome c | 4.43 ± 0.73 |
| BSA—purified fraction | 15.04 ± 1.44 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zámocký, M.; Kamlárová, A.; Maresch, D.; Chovanová, K.; Harichová, J.; Furtmüller, P.G. Hybrid Heme Peroxidases from Rice Blast Fungus Magnaporthe oryzae Involved in Defence against Oxidative Stress. Antioxidants 2020, 9, 655. https://doi.org/10.3390/antiox9080655
Zámocký M, Kamlárová A, Maresch D, Chovanová K, Harichová J, Furtmüller PG. Hybrid Heme Peroxidases from Rice Blast Fungus Magnaporthe oryzae Involved in Defence against Oxidative Stress. Antioxidants. 2020; 9(8):655. https://doi.org/10.3390/antiox9080655
Chicago/Turabian StyleZámocký, Marcel, Anna Kamlárová, Daniel Maresch, Katarína Chovanová, Jana Harichová, and Paul G. Furtmüller. 2020. "Hybrid Heme Peroxidases from Rice Blast Fungus Magnaporthe oryzae Involved in Defence against Oxidative Stress" Antioxidants 9, no. 8: 655. https://doi.org/10.3390/antiox9080655
APA StyleZámocký, M., Kamlárová, A., Maresch, D., Chovanová, K., Harichová, J., & Furtmüller, P. G. (2020). Hybrid Heme Peroxidases from Rice Blast Fungus Magnaporthe oryzae Involved in Defence against Oxidative Stress. Antioxidants, 9(8), 655. https://doi.org/10.3390/antiox9080655