Super-Resolution Imaging of Plant Receptor-Like Kinases Uncovers Their Colocalization and Coordination with Nanometer Resolution
Abstract
:1. Introduction
2. Methods
2.1. Materials
2.2. Construction of Transgenic Plants
2.3. Arabidopsis Growth
2.4. Immunostaining
2.5. Fluorescence Microscopy
2.6. Image Processing
3. Results
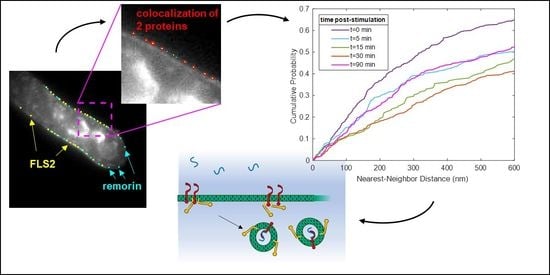
3.1. Quantitative Analysis of FLS2 and Remorin Colocalization in the Membrane at Increasing Time Points following Ligand Stimulation
3.2. Quantitative Analysis of P2K1 and CERK1 Colocalization with FLS2 or Remorin
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tena, G.; Boudsocq, M.; Sheen, J. Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 2011, 14, 519–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, D.; Wang, G.; Zhou, J.M. Receptor kinases in plant-pathogen interactions: More than pattern recognition. Plant Cell 2017, 29, 618–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, D.; Kang, R.; Coyn, A.B.; Zeh, H.J.; Lotze, M.T. PAMPs and DAMPs: Signal 0s that spur autophagy and immunity. Immunol. Rev. 2012, 249, 158–175. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhao, F.; Jiang, L.; Chen, C.; Wu, L.; Liu, Z. Different pathogen defense strategies in arabidopsis: More than pathogen recognition. Cells 2018, 7, 252. [Google Scholar] [CrossRef] [Green Version]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–407. [Google Scholar] [CrossRef]
- Martiniere, A.; Zelazny, E. Membrane nanodomains and transport functions in plant. Plant Physiol. 2021, 187, 1839–1855. [Google Scholar] [CrossRef]
- Ott, T. Membrane nanodomains and microdomains in plant–microbe interactions. Curr. Opin. Plant Biol. 2017, 40, 82–88. [Google Scholar] [CrossRef]
- Jaillais, Y.; Ott, T. The nanoscale organization of the plasma membrane and its importance in signaling: A proteolipid perspective. Plant Physiol. 2020, 182, 1682–1696. [Google Scholar] [CrossRef] [Green Version]
- Gronnier, J.; Gerbeau-Pissot, P.; Germain, V.; Mongrand, S.; Simon-Plas, F. Divide and Rule: Plant Plasma Membrane Organization. Trends Plant Sci. 2018, 23, 899–917. [Google Scholar] [CrossRef]
- Liang, P.; Stratil, T.F.; Popp, C.; Marín, M.; Folgmann, J.; Mysore, K.S.; Wen, J.; Ott, T. Symbiotic root infections in Medicago truncatula require remorin-mediated receptor stabilization in membrane nanodomains. Proc. Natl. Acad. Sci. USA 2018, 115, 5289–5294. [Google Scholar] [CrossRef]
- Haney, C.H.; Riely, B.K.; Tricoli, D.M.; Cook, D.R.; Ehrhardt, D.W.; Long, S.R. Symbiotic rhizobia bacteria trigger a change in localization and dynamics of the Medicago truncatula receptor kinase LYK3. Plant Cell 2011, 23, 2774–2787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gui, J.; Zheng, S.; Liu, C.; Shen, J.; Li, J.; Li, L. OsREM4.1 Interacts with OsSERK1 to Coordinate the Interlinking between Abscisic Acid and Brassinosteroid Signaling in Rice. Dev. Cell 2016, 38, 201–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raffaele, S.; Bayer, E.; Lafarge, D.; Cluzet, S.; Retana, S.G.; Boubekeur, T.; Castel, N.L.; Carde, J.P.; Lherminier, J.; Noirot, E.; et al. Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell 2009, 21, 1541–1555. [Google Scholar] [CrossRef] [Green Version]
- Mongrand, S.; Morel, J.; Laroche, J.; Claverol, S.; Carde, J.P.; Hartmann, M.A.; Bonneu, M.; Simon-Plas, F.; Lessire, R.; Bessoule, J.J. Lipid rafts in higher plant cells: Purification and characterization of triton X-100-insoluble microdomains from tobacco plasma membrane. J. Biol. Chem. 2004, 279, 36277–36286. [Google Scholar] [CrossRef] [Green Version]
- Bücherl, C.A.; Jarsch, I.K.; Schudoma, C.; Segonzac, C.; Mbengue, M.; Robatze, S.; MacLean, D.; Ott, T.; Zipfe, C. Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. eLife 2017, 6, e25114. [Google Scholar] [CrossRef]
- Huang, D.; Sun, Y.; Ma, Z.; Ke, M.; Cui, Y.; Chen, Z.; Chen, C.; Ji, C.; Tran, T.M.; Yang, L.; et al. Salicylic acid-mediated plasmodesmal closure via Remorin-dependent lipid organization. Proc. Natl. Acad. Sci. USA 2019, 116, 21274–21284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Li, H.; Lv, X.; Chen, T.; Li, R.; Xue, Y.; Jiang, J.; Jin, B.; Baluška, F.; Šamaj, J.; et al. Spatiotemporal dynamics of the BRI1 receptor and its regulation by membrane microdomains in living Arabidopsis cells. Mol. Plant 2015, 8, 1334–1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, B.; Zhang, X.; Li, L.; Abbas, S.; Yu, M.; Cui, Y.; Baluška, F.; Hwang, I.; Shan, X.; Lin, J. Dynamic spatial reorganization of BSK1 complexes in the plasma membrane underpins signal-specific activation for growth and immunity. Mol. Plant 2021, 14, 588–603. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Ahsan, N.; Thelen, J.J.; Stacey, G. S-Acylation of plant immune receptors mediates immune signaling in plasma membrane nanodomains. bioRxiv 2019, 720482. [Google Scholar] [CrossRef] [Green Version]
- Millet, Y.A.; Danna, C.H.; Clay, N.K.; Songnuan, W.; Simon, M.D.; Werck-Reichhart, D.; Ausubel, F.M. Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 2010, 22, 973–990. [Google Scholar] [CrossRef]
- Robatzek, S.; Chinchilla, D.; Boller, T. Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 2006, 20, 537–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.; Tanaka, K.; Cao, Y.; Qi, Y.; Qiu, J.; Liang, Y.; Lee, S.Y.; Stacey, G. Identification of a plant receptor for extracellular ATP. Science 2014, 343, 290–294. [Google Scholar] [CrossRef]
- Cao, Y.; Liang, Y.; Tanaka, K.; Nguyen, C.T.; Jedrzejczak, R.P.; Joachimiak, A.; Stacey, G. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 2014, 3, e03766. [Google Scholar] [CrossRef] [PubMed]
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 19613–19618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denoux, C.; Galletti, R.; Mammarella, N.; Gopalan, S.; Werck, D.; De Lorenzo, G.; Ferrari, S.; Ausubel, F.M.; Dewdney, J. Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol. Plant 2008, 1, 423–445. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Kim, P.; Yu, L.; Cai, G.; Chen, S.; Alfano, J.R.; Zhou, J.M. Activation-Dependent Destruction of a Co-receptor by a Pseudomonas syringae Effector Dampens Plant Immunity. Cell Host Microbe 2016, 20, 504–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matern, A.; Böttcher, C.; Eschen-Lippold, L.; Westermann, B.; Smolka, U.; Döll, S.; Trempel, F.; Aryal, B.; Scheel, D.; Geisler, M.; et al. A substrate of the ABC transporter PEN3 stimulates bacterial flagellin (flg22)-induced callose deposition in Arabidopsis thaliana. J. Biol. Chem. 2019, 294, 6857–6870. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Yamaguchi, K.; Shirakawa, T.; Nakagami, H.; Mine, A.; Ishikawa, K.; Fujiwara, M.; Narusaka, M.; Narusaka, Y.; Ichimura, K.; et al. The Arabidopsis CERK 1-associated kinase PBL 27 connects chitin perception to MAPK activation. EMBO J. 2016, 35, 2468–2483. [Google Scholar] [CrossRef]
- Thor, K.; Peiter, E. Cytosolic calcium signals elicited by the pathogen-associated molecular pattern flg22 in stomatal guard cells are of an oscillatory nature. New Phytol. 2014, 204, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Monaghan, J.; Matschi, S.; Romeis, T.; Zipfel, C. The calcium-dependent protein kinase CPK28 negatively regulates the BIK1-mediated PAMPinduced calcium burst. Plant Signal. Behav. 2015, 10, e1018497. [Google Scholar] [CrossRef]
- Chen, D.; Hao, F.; Mu, H.; Ahsan, N.; Thelen, J.J.; Stacey, G. S-acylation of P2K1 mediates extracellular ATP-induced immune signaling in Arabidopsis. Nat. Commun. 2021, 12, 2750. [Google Scholar] [CrossRef] [PubMed]
- Gimenez-Ibanez, S.; Ntoukakis, V.; Rathjen, J.P. The LysM receptor kinase CERK1 mediates bacterial perception in Arabidopsis. Plant Signal. Behav. 2009, 4, 539–541. [Google Scholar] [CrossRef] [PubMed]
- Espinoza, C.; Liang, Y.; Stacey, G. Chitin receptor CERK1 links salt stress and chitin-triggered innate immunity in Arabidopsis. Plant J. 2017, 89, 984–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De, I.; Sadhukhan, S. Emerging Roles of DHHC-mediated Protein S-palmitoylation in Physiological and Pathophysiological Context. Eur. J. Cell Biol. 2018, 97, 319–338. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.W.; Niu, Y.; Jia, Y.; Ordon, J.; Copeland, C.; Emonet, A.; Geldner, N.; Guan, R.; Stolze, S.C.; Nakagami, H.; et al. Coordination of microbe–host homeostasis by crosstalk with plant innate immunity. Nat. Plants 2021, 7, 814–825. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Liu, J.; Wang, J.; Chen, S.; Chen, L.; Wang, J.; Bin Wang, H.; Liu, B. The juxtamembrane domains of Arabidopsis CERK1, BAK1, and FLS2 play a conserved role in chitin-induced signaling. J. Integr. Plant Biol. 2020, 62, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.M.; Chng, C.P.; Pu, X.; Ma, Z.; Han, X.; Liu, X.; Yang, L.; Huang, C.; Miao, Y. Potentiation of plant defense by bacterial outer membrane vesicles is mediated by membrane nanodomains. Plant Cell 2022, 34, 395–418. [Google Scholar] [CrossRef]
- Mbengue, M.; Bourdais, G.; Gervasi, F.; Beck, M.; Zhou, J.; Spallek, T.; Bartels, S.; Boller, T.; Ueda, T.; Kuhn, H.; et al. Clathrin-dependent endocytosis is required for immunity mediated by pattern recognition receptor kinases. Proc. Natl. Acad. Sci. USA 2016, 113, 11034–11039. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Claus, L.A.N.; Leslie, M.E.; Tao, K.; Wu, Z.; Liu, J.; Yu, X.; Li, B.; Zhou, J.; Savatin, D.V.; et al. Ligand-induced monoubiquitination of BIK1 regulates plant immunity. Nature 2020, 581, 199–203. [Google Scholar] [CrossRef]
- Srivastava, M.; Srivastava, A.K.; Orosa-Puente, B.; Campanaro, A.; Zhang, C.; Sadanandom, A. SUMO Conjugation to BZR1 Enables Brassinosteroid Signaling to Integrate Environmental Cues to Shape Plant Growth. Curr. Biol. 2020, 30, 1410–1423. [Google Scholar] [CrossRef]
- Henriques, R.; Griffiths, C.; Rego, E.H.; Mhlanga, M.M. PALM and STORM: Unlocking live-cell super-resolution. Biopolymers 2011, 95, 322–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, D.; Zhao, B.; Xie, Y.; Orr, G.; Li, A.D.Q. Understanding super-resolution nanoscopy and its biological applications in cell imaging. Phys. Chem. Chem. Phys. 2013, 15, 14856–14861. [Google Scholar] [CrossRef]
- Jarsch, I.K.; Konrad, S.S.A.; Stratil, T.F.; Urbanus, S.L.; Szymanski, W.; Braun, P.; Braun, K.H.; Ott, T. Plasma membranes are Subcompartmentalized into a plethora of coexisting and diverse microdomains in Arabidopsis and Nicotiana benthamiana. Plant Cell 2014, 26, 1698–1711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Hu, D.; Dieter, C.; Ansong, C.; Sussel, L.; Orr, G. Single molecule-based fliFISH validates radial and heterogeneous gene expression patterns in pancreatic islet β cells. Diabetes 2021, 70, 1117–1122. [Google Scholar] [CrossRef]
- Li, F.; Mitchell, H.D.; Mensch, A.C.; Hu, D.; Laudadio, E.D.; Hedlund Orbeck, J.K.; Hamers, R.J.; Orr, G. Expression Patterns of Energy-Related Genes in Single Cells Uncover Key Isoforms and Enzymes That Gain Priority under Nanoparticle-Induced Stress. ACS Nano 2021, 16, 7197–7209. [Google Scholar] [CrossRef]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.M.; He, S.Y.; Xin, X.F. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef]
- Roux, M.E.; Rasmussen, M.W.; Palma, K.; Lolle, S.; Regué, À.M.; Bethke, G.; Glazebrook, J.; Zhang, W.; Sieburth, L.; Larsen, M.R.; et al. The mRNA decay factor PAT 1 functions in a pathway including MAP kinase 4 and immune receptor SUMM 2. EMBO J. 2015, 34, 593–608. [Google Scholar] [CrossRef] [Green Version]
- Kleine-Vehn, J.; Wabnik, K.; Martinière, A.; Łangowski, Ł.; Willig, K.; Naramoto, S.; Leitner, J.; Tanaka, H.; Jakobs, S.; Robert, S.; et al. Recycling, clustering, and endocytosis jointly maintain PIN auxin carrier polarity at the plasma membrane. Mol. Syst. Biol. 2011, 7, 540. [Google Scholar] [CrossRef]
- McKenna, J.F.; Rolfe, D.J.; Webb, S.E.D.; Tolmie, A.F.; Botchway, S.W.; Martin-Fernandez, M.L.; Hawes, C.; Runions, J. The cell wall regulates dynamics and size of plasma-membrane nanodomains in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 12857–12862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demir, F.; Horntrich, C.; Blachutzik, J.O.; Scherzer, S.; Reinders, Y.; Kierszniowska, S.; Schulze, W.X.; Harms, G.S.; Hedrich, R.; Geiger, D.; et al. Arabidopsis nanodomain-delimited ABA signaling pathway regulates the anion channel SLAH3. Proc. Natl. Acad. Sci. USA 2013, 110, 8296–8301. [Google Scholar] [CrossRef] [Green Version]
- Gronnier, J.; Franck, C.M.; Stegmann, M.; Defalco, T.A.; Cifuentes, A.A.; Dünser, K.; Lin, W.; Yang, Z.; Kleine-Vehn, J.; Ringli, C.; et al. FERONIA regulates FLS2 plasma membrane nanoscale dynamics to modulate plant immune signaling. bioRxiv 2020. [Google Scholar] [CrossRef]
- Cui, Y.; Li, X.; Yu, M.; Li, R.; Fan, L.; Zhu, Y.; Lin, J. Sterols regulate endocytic pathways during flg22-induced defense responses in arabidopsis. Development 2018, 145, dev165688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danna, C.H.; Millet, Y.A.; Koller, T.; Han, S.W.; Bent, A.F.; Ronald, P.C.; Ausubel, F.M. The Arabidopsis flagellin receptor FLS2 mediates the perception of Xanthomonas Ax21 secreted peptides. Proc. Natl. Acad. Sci. USA 2011, 108, 9286–9291. [Google Scholar] [CrossRef] [Green Version]
- Smith, J.M.; Salamango, D.J.; Leslie, M.E.; Collins, C.A.; Heese, A. Sensitivity to Flg22 Is modulated by ligand-induced degradation and de novo synthesis of the endogenous flagellin-receptor FLAGELLIN-SENSING2. Plant Physiol. 2014, 164, 440–454. [Google Scholar] [CrossRef] [Green Version]
- Jeworutzki, E.; Roelfsema, M.R.G.; Anschütz, U.; Krol, E.; Elzenga, J.T.M.; Felix, G.; Boller, T.; Hedrich, R.; Becker, D. Early signaling through the arabidopsis pattern recognition receptors FLS2 and EFR involves Ca2+-associated opening of plasma membrane anion channels. Plant J. 2010, 62, 367–378. [Google Scholar] [CrossRef]
- Keinath, N.F.; Kierszniowska, S.; Lorek, J.; Bourdais, G.; Kessler, S.A.; Shimosato-Asano, H.; Grossniklaus, U.; Schulze, W.X.; Robatzek, S.; Panstruga, R. PAMP (Pathogen-associated Molecular Pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J. Biol. Chem. 2010, 285, 39140–39149. [Google Scholar] [CrossRef] [Green Version]
- Tran, T.M.; Ma, Z.; Triebl, A.; Nath, S.; Cheng, Y.; Gong, B.Q.; Han, X.; Wang, J.; Li, J.F.; Wenk, M.R.; et al. The bacterial quorum sensing signal DSF hijacks Arabidopsis thaliana sterol biosynthesis to suppress plant innate immunity. Life Sci. Alliance 2020, 3, 10. [Google Scholar] [CrossRef]
- Beck, M.; Zhou, J.; Faulkner, C.; Mac, D.L.; Robatzek, S. Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell 2012, 24, 4205–4219. [Google Scholar] [CrossRef]
- Lefebvre, B.; Timmers, T.; Mbengue, M.; Moreau, S.; Hervé, C.; Tóth, K.; Bittencourt-Silvestre, J.; Klaus, D.; Deslandes, L.; Godiard, L.; et al. A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc. Natl. Acad. Sci. USA 2010, 107, 2343–2348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, K.; Waletich, J.R.; Arredondo, F.; Tyler, B.M. Manipulating endoplasmic reticulum-plasma membrane tethering in plants through fluorescent protein complementation. Front. Plant Sci. 2019, 10, 635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bozkurt, T.O.; Belhaj, K.; Dagdas, Y.F.; Chaparro-Garcia, A.; Wu, C.H.; Cano, L.M.; Kamoun, S. Rerouting of Plant Late Endocytic Trafficking Toward a Pathogen Interface. Traffic 2015, 16, 204–226. [Google Scholar] [CrossRef] [PubMed]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Traeger, J.; Hu, D.; Yang, M.; Stacey, G.; Orr, G. Super-Resolution Imaging of Plant Receptor-Like Kinases Uncovers Their Colocalization and Coordination with Nanometer Resolution. Membranes 2023, 13, 142. https://doi.org/10.3390/membranes13020142
Traeger J, Hu D, Yang M, Stacey G, Orr G. Super-Resolution Imaging of Plant Receptor-Like Kinases Uncovers Their Colocalization and Coordination with Nanometer Resolution. Membranes. 2023; 13(2):142. https://doi.org/10.3390/membranes13020142
Chicago/Turabian StyleTraeger, Jeremiah, Dehong Hu, Mengran Yang, Gary Stacey, and Galya Orr. 2023. "Super-Resolution Imaging of Plant Receptor-Like Kinases Uncovers Their Colocalization and Coordination with Nanometer Resolution" Membranes 13, no. 2: 142. https://doi.org/10.3390/membranes13020142
APA StyleTraeger, J., Hu, D., Yang, M., Stacey, G., & Orr, G. (2023). Super-Resolution Imaging of Plant Receptor-Like Kinases Uncovers Their Colocalization and Coordination with Nanometer Resolution. Membranes, 13(2), 142. https://doi.org/10.3390/membranes13020142