The Implant Proteome—The Right Surgical Glue to Fix Titanium Implants In Situ
Abstract
:1. Introduction
2. Materials and Methods
2.1. Implants
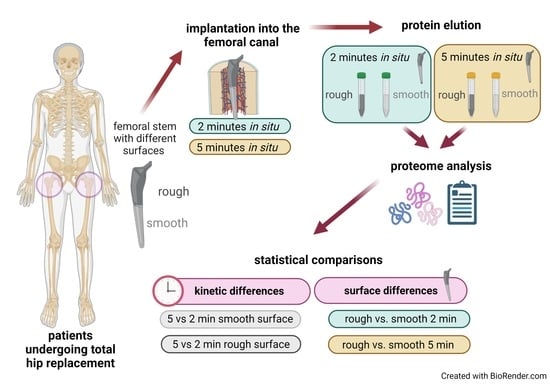
2.2. Probands, Surgical Technique and Implant Retrieval
2.3. Protein Quantification
2.4. Proteome Analysis
2.5. Annotation of Proteomics Data and Bioinformatics Analysis
3. Results
3.1. Patients Characteristic
3.2. Characterization of the In Situ Implant Proteome
3.3. Changes in Protein Adsorption on Titanium Implants
3.3.1. Proteomic Differences between 2 and 5 Min In Situ
3.3.2. Proteomic Differences between the Smooth and the Rough Surfaces
4. Discussion
4.1. Proteins of the Bone ECM and Their Distribution within the Implant Proteome
4.2. Hemostasis and Inflammation
4.2.1. Proteins Potentially Involved in Hemostasis and Neutrophil Activity
4.2.2. Possible Neutrophil Recruitment through DAMPs and the Complement System
4.3. Limitations of Our Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van Oldenrijk, J.; Scholtes, V.A.B.; van Beers, L.W.A.H.; Geerdink, C.H.; Niers, B.B.A.M.; Runne, W.; Bhandari, M.; Poolman, R.W. Better early functional outcome after short stem total hip arthroplasty? A prospective blinded randomised controlled multicentre trial comparing the Collum Femoris Preserving stem with a Zweymuller straight cementless stem total hip replacement for the treatment of primary osteoarthritis of the hip. BMJ Open 2017, 7, e014522. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, M.; Jennissen, H.P.; Haversath, M.; Busch, A.; Grupp, T.; Sowislok, A.; Herten, M. Intrasurgical Protein Layer on Titanium Arthroplasty Explants: From the Big Twelve to the Implant Proteome. Proteom. Clin. Appl. 2019, 13, e1800168. [Google Scholar] [CrossRef] [PubMed]
- Sowislok, A.; Weischer, T.; Jennissen, H.P. The In Situ Human Dental Implantome: A First Appraisal. Curr. Dir.Biomed. Eng. 2021, 7, 827–830. [Google Scholar] [CrossRef]
- Busch, A.; Jäger, M.; Mayer, C.; Sowislok, A. Functionalization of Synthetic Bone Substitutes. Int. J. Mol. Sci. 2021, 22, 4412. [Google Scholar] [CrossRef] [PubMed]
- Abaricia, J.O.; Shah, A.H.; Musselman, R.M.; Olivares-Navarrete, R. Hydrophilic titanium surfaces reduce neutrophil inflammatory response and NETosis. Biomater. Sci. 2020, 8, 2289–2299. [Google Scholar] [CrossRef] [Green Version]
- Vroman, L.; Adams, A.L. Findings with the recording ellipsometer suggesting rapid exchange of specific plasma proteins at liquid/solid interfaces. Surf. Sci. 1969, 16, 438–446. [Google Scholar] [CrossRef]
- Vroman, L.; Adams, A.L. Identification of rapid changes at plasma-solid interfaces. J. Biomed. Mater. Res. 1969, 3, 43–67. [Google Scholar] [CrossRef]
- Swamy, G.; Pace, A.; Quah, C.; Howard, P. The Bicontact cementless primary total hip arthroplasty: Long-term results. Int. Orthop. 2012, 36, 915–920. [Google Scholar] [CrossRef] [Green Version]
- Ateschrang, A.; Weise, K.; Weller, S.; Stöckle, U.; Zwart, P.D.; Ochs, B.G. Long-term results using the straight tapered femoral cementless hip stem in total hip arthroplasty: A minimum of twenty-year follow-up. J. Arthroplast. 2014, 29, 1559–1565. [Google Scholar] [CrossRef]
- Learmonth, I.D. (Ed.) Interfaces in Total Hip Arthroplasty; Springer: London, UK, 2000. [Google Scholar]
- Bauer, R.; Kerschbaumer, F.; Poisel, S.; Oberthaler, W. The transgluteal approach to the hip joint. Arch. Orthop. Trauma. Surg. 1979, 95, 47–49. [Google Scholar] [CrossRef]
- Hardinge, K. The direct lateral approach to the hip. J. Bone Joint Surg. Br. 1982, 64, 17–19. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.; Rosebrough, N.; Farr, A.L.; Randall, R. Protein Measurement with the Folin Phenol Reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Hill, H.D.; Straka, J.G. Protein Determination Using Bicinchoninic Acid in the Presence of Sulfhydryl Reagents. Anal. Biochem. 1988, 170, 203–208. [Google Scholar] [CrossRef]
- Makridakis, M.; Vlahou, A. GeLC-MS: A Sample Preparation Method for Proteomics Analysis of Minimal Amount of Tissue. Methods Mol. Biol. 2018, 1788, 165–175. [Google Scholar] [CrossRef]
- Lygirou, V.; Latosinska, A.; Makridakis, M.; Mullen, W.; Delles, C.; Schanstra, J.P.; Zoidakis, J.; Pieske, B.; Mischak, H.; Vlahou, A. Plasma proteomic analysis reveals altered protein abundances in cardiovascular disease. J. Transl. Med. 2018, 16, 104. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Dakna, M.; Harris, K.; Kalousis, A.; Carpentier, S.; Kolch, W.; Schanstra, J.P.; Haubitz, M.; Vlahou, A.; Mischak, H.; Girolami, M. Addressing the challenge of defining valid proteomic biomarkers and classifiers. BMC Bioinform. 2010, 11, 594. [Google Scholar] [CrossRef] [Green Version]
- Nanjappa, V.; Thomas, J.K.; Marimuthu, A.; Muthusamy, B.; Radhakrishnan, A.; Sharma, R.; Ahmad Khan, A.; Balakrishnan, L.; Sahasrabuddhe, N.A.; Kumar, S.; et al. Plasma Proteome Database as a resource for proteomics research: 2014 update. Nucleic Acids Res. 2014, 42, D959–D965. [Google Scholar] [CrossRef] [Green Version]
- Farrah, T.; Deutsch, E.W.; Omenn, G.S.; Campbell, D.S.; Sun, Z.; Bletz, J.A.; Mallick, P.; Katz, J.E.; Malmström, J.; Ossola, R.; et al. A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol. Cell. Proteom. 2011, 10, M110.006353. [Google Scholar] [CrossRef] [Green Version]
- Anderson, N.L.; Polanski, M.; Pieper, R.; Gatlin, T.; Tirumalai, R.S.; Conrads, T.P.; Veenstra, T.D.; Adkins, J.N.; Pounds, J.G.; Fagan, R.; et al. The human plasma proteome: A nonredundant list developed by combination of four separate sources. Mol. Cell. Proteom. 2004, 3, 311–326. [Google Scholar] [CrossRef] [Green Version]
- Geyer, P.E.; Kulak, N.A.; Pichler, G.; Holdt, L.M.; Teupser, D.; Mann, M. Plasma Proteome Profiling to Assess Human Health and Disease. Cell Syst. 2016, 2, 185–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bryk, A.H.; Wiśniewski, J.R. Quantitative Analysis of Human Red Blood Cell Proteome. J. Proteome Res. 2017, 16, 2752–2761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Human Protein Atlas. Available online: https://www.proteinatlas.org (accessed on 16 December 2021).
- Shao, X.; Taha, I.N.; Clauser, K.R.; Gao, Y.T.; Naba, A. MatrisomeDB: The ECM-protein knowledge database. Nucleic Acids Res. 2020, 48, D1136–D1144. [Google Scholar] [CrossRef] [PubMed]
- MatrisomeDB. Available online: http://matrisomedb.pepchem.org (accessed on 16 December 2021).
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- ShinyGO. Available online: http://bioinformatics.sdstate.edu/go/ (accessed on 16 December 2021).
- Brånemark, P.I. Osseointegration and its experimental background. J. Prosthet. Dent. 1983, 50, 399–410. [Google Scholar] [CrossRef]
- Mavrogenis, A.F.; Dimitriou, R.; Parvizi, J.; Babis, G.C. Biology of implant osseointegration. J. Musculoskelet Neuronal Interact 2009, 9, 61–71. [Google Scholar]
- Robert, A.; Latour, J.R. Biomaterials: Protein—Surface Interactions. In Encyclopedia of Biomaterials and Biomedical Engineering, 2nd ed.; Gary, E., Gary, W., Bowlin, L., Eds.; CRC Press: Boca Raton, FL, USA, 2008; pp. 270–284. ISBN 9781420078022. [Google Scholar]
- Mendonça, D.B.S.; Miguez, P.A.; Mendonça, G.; Yamauchi, M.; Aragão, F.J.L.; Cooper, L.F. Titanium surface topography affects collagen biosynthesis of adherent cells. Bone 2011, 49, 463–472. [Google Scholar] [CrossRef]
- Lin, X.; Patil, S.; Gao, Y.-G.; Qian, A. The Bone Extracellular Matrix in Bone Formation and Regeneration. Front. Pharmacol. 2020, 11, 757. [Google Scholar] [CrossRef]
- Gehron Robey, P.G. Robey. Bone Matrix Proteoglycans and Glycoproteins. In Principles of Bone Biology, 2nd ed.; Bilezikian, J., Raisz, L., Rodan, G., Eds.; Academic Press: San Diego, CA, USA, 2002; pp. 225–237. [Google Scholar]
- Sroga, G.E.; Vashishth, D. Effects of bone matrix proteins on fracture and fragility in osteoporosis. Curr. Osteoporos. Rep. 2012, 10, 141–150. [Google Scholar] [CrossRef] [Green Version]
- Thalji, G.N.; Nares, S.; Cooper, L.F. Early molecular assessment of osseointegration in humans. Clin. Oral Implant. Res. 2014, 25, 1273–1285. [Google Scholar] [CrossRef]
- Albrektsson, T.; Dahlin, C.; Jemt, T.; Sennerby, L.; Turri, A.; Wennerberg, A. Is marginal bone loss around oral implants the result of a provoked foreign body reaction? Clin. Implant Dent. Relat. Res. 2014, 16, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Trindade, R.; Albrektsson, T.; Tengvall, P.; Wennerberg, A. Foreign Body Reaction to Biomaterials: On Mechanisms for Buildup and Breakdown of Osseointegration. Clin. Implant Dent. Relat. Res. 2016, 18, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Shiu, H.T.; Goss, B.; Lutton, C.; Crawford, R.; Xiao, Y. Formation of blood clot on biomaterial implants influences bone healing. Tissue Eng. Part B Rev. 2014, 20, 697–712. [Google Scholar] [CrossRef]
- Teller, P.; White, T.K. The Physiology of Wound Healing: Injury Through Maturation. Perioper. Nurs. Clin. 2011, 6, 159–170. [Google Scholar] [CrossRef]
- Golebiewska, E.M.; Poole, A.W. Platelet secretion: From haemostasis to wound healing and beyond. Blood Rev. 2015, 29, 153–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zucoloto, A.Z.; Jenne, C.N. Platelet-Neutrophil Interplay: Insights Into Neutrophil Extracellular Trap (NET)-Driven Coagulation in Infection. Front. Cardiovasc. Med. 2019, 6, 85. [Google Scholar] [CrossRef] [Green Version]
- Trindade, R.; Albrektsson, T.; Galli, S.; Prgomet, Z.; Tengvall, P.; Wennerberg, A. Osseointegration and foreign body reaction: Titanium implants activate the immune system and suppress bone resorption during the first 4 weeks after implantation. Clin. Implant Dent. Relat. Res. 2018, 20, 82–91. [Google Scholar] [CrossRef]
- El Kholy, K.; Buser, D.; Wittneben, J.-G.; Bosshardt, D.D.; van Dyke, T.E.; Kowolik, M.J. Investigating the Response of Human Neutrophils to Hydrophilic and Hydrophobic Micro-Rough Titanium Surfaces. Materials 2020, 13, 3421. [Google Scholar] [CrossRef]
- Lacy, P. Mechanisms of Degranulation in Neutrophils. Allergy Asthma Clin. Immunol. 2006, 2, 98–108. [Google Scholar] [CrossRef] [Green Version]
- Aratani, Y. Myeloperoxidase: Its role for host defense, inflammation, and neutrophil function. Arch. Biochem. Biophys. 2018, 640, 47–52. [Google Scholar] [CrossRef]
- Zamolodchikova, T.S.; Tolpygo, S.M.; Svirshchevskaya, E.V. Cathepsin G-Not Only Inflammation: The Immune Protease Can Regulate Normal Physiological Processes. Front. Immunol. 2020, 11, 411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pittman, K.; Kubes, P. Damage-associated molecular patterns control neutrophil recruitment. J. Innate Immun. 2013, 5, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. Neutrophils in tissue injury and repair. Cell Tissue Res. 2018, 371, 531–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryckman, C.; Vandal, K.; Rouleau, P.; Talbot, M.; Tessier, P.A. Proinflammatory activities of S100: Proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. J. Immunol. 2003, 170, 3233–3242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mödinger, Y.; Teixeira, G.Q.; Neidlinger-Wilke, C.; Ignatius, A. Role of the Complement System in the Response to Orthopedic Biomaterials. Int. J. Mol. Sci. 2018, 19, 3367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoengraf, P.; Lambris, J.D.; Recknagel, S.; Kreja, L.; Liedert, A.; Brenner, R.E.; Huber-Lang, M.; Ignatius, A. Does complement play a role in bone development and regeneration? Immunobiology 2013, 218, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Campos, V.; Melo RC, N.; Silva, L.P.; Aquino, E.N.; Castro, M.S.; Fontes, W. Characterization of neutrophil adhesion to different titanium surfaces. Bull. Mater. Sci. 2014, 37, 157–166. [Google Scholar] [CrossRef] [Green Version]
- Dayer, R.; Brennan, T.C.; Rizzoli, R.; Ammann, P. PTH improves titanium implant fixation more than pamidronate or renutrition in osteopenic rats chronically fed a low protein diet. Osteoporos. Int. 2010, 21, 957–967. [Google Scholar] [CrossRef] [Green Version]



| Variable | Value | |
|---|---|---|
| Number of patients | 12 | |
| Age (years) | 73.5 ± 7.34 | |
| Male | 3 (25%) | |
| BMI | 27.6 ± 3.01 | |
| Surgery site | left | 5 (42%) |
| right | 7 (58%) | |
| Surgery time (min) | 88.5 ± 7.56 | |
| Hospital time (days) | 11.4 ± 3.18 | |
| Comorbidities | ||
| Hypertension | 6 (50%) | |
| ACVB (aorto-coronary-vein-bypass) | 2 (16.6%) | |
| Hypothyreosis | 2 (16.6%) | |
| Rheumatoid arthritis | 2 (16.6%) | |
| Adipositas | 1 (8.3%) | |
| Diabetes mellitus | 1 (8.3%) | |
| Variable | Time Point | Mean | Standard Deviation | Normal Values |
|---|---|---|---|---|
| Sodium (mmol/L) | −1 | 139.8 | 4.2 | |
| 0 | 138.9 | 3.7 | 136–145 | |
| 2 | 138.4 | 3.7 | ||
| Potassium (mmol/L) | −1 | 4.6 | 0.3 | |
| 0 | 4.2 | 0.3 | 3.5–5.1 | |
| 2 | 4.0 | 0.3 | ||
| Leucocytes (1/nL) | −1 | 7.2 | 1.7 | |
| 0 | 11.8 | 2.8 | 4.0–11.0 | |
| 2 | 9.6 | 2.9 | ||
| Hemoglobin (g/dL) | −1 | 14.0 | 1.2 | |
| 0 | 11.8 | 1.9 | 11.6–16.3 | |
| 2 | 10.2 | 1.5 | ||
| Thrombocytes (1/nL) | −1 | 255.8 | 63.3 | |
| 0 | 213.7 | 38.2 | 140–320 | |
| 2 | 195.2 | 40.5 | ||
| Orotein (g/L) | −1 | 67.1 | 5.9 | |
| 0 | 58.3 | 8.1 | 64–83 | |
| 2 | 54.0 | 3.4 | ||
| Quick (%) | −1 | 101.4 | 23.9 | |
| 0 | 91.0 | 12.9 | 70–130 | |
| 2 | 96.5 | 16.8 | ||
| pTT (s) | −1 | 26.6 | 3.8 | |
| 0 | 26.1 | 2.4 | 24–32.2 | |
| −1 | 28.2 | 1.9 | ||
| Fibrinogen (mg/dL) | −1 | 324.3 | 66.2 | |
| 0 | 312.8 | 63.8 | 180–350 | |
| 2 | 584.6 | 196.4 |
| Surface/Condition | Sample Volume (ml) | Protein Concentration (µg/mL) | Total Protein (µg) |
|---|---|---|---|
| Rough/2 min | 3.5 ± 0.8 | 66.0 ± 30.0 | 231.2 ± 133.0 |
| Rough/5 min | 3.2 ± 0.3 | 69.2 ± 25.0 | 221.3 ± 93.5 |
| Smooth/2 min | 3.4 ± 0.5 | 13.3 ± 8.6 | 40.9 ± 18.6 |
| Smooth/5 min | 2.9 ± 0.6 | 5.1 ± 2.4 | 15.3 ± 8.6 |
| Localization | Name | # Peptides | Avg. Abund. |
|---|---|---|---|
| Intracellular/Membrane | Hemoglobin subunit beta | 25 | 139,291.93 |
| Hemoglobin subunit alpha | 25 | 42,991.18 | |
| Protein AHNAK2 | 10 | 5187.38 | |
| Keratin, type II cytoskeletal 1 | 21 | 4755.28 | |
| Hemoglobin subunit delta | 12 | 4421.55 | |
| Actin, cytoplasmic 1 | 16 | 3895.84 | |
| Carbonic anhydrase 1 | 19 | 3776.61 | |
| Spectrin beta chain, erythrocytic | 40 | 2911.55 | |
| Myosin-7 | 99 | 2564.4 | |
| Peroxiredoxin-2 | 16 | 2186.3 | |
| Secreted to blood | Serum albumin | 99 | 70,046.62 |
| Alpha-1-antichymotrypsin | 12 | 7424.58 | |
| Fibrinogen beta chain | 28 | 2523.37 | |
| Alpha-1-antitrypsin | 27 | 2361.18 | |
| Fibrinogen gamma chain | 26 | 2286.78 | |
| Serotransferrin | 49 | 2063.98 | |
| Alpha-2-macroglobulin | 56 | 1660.91 | |
| Apolipoprotein B-100 | 98 | 1433.53 | |
| Fibrinogen alpha chain | 29 | 1398.07 | |
| Complement C3 | 85 | 1241.86 | |
| ECM | Collagen alpha-1(II) chain | 67 | 6790.18 |
| Collagen alpha-1(XXIV) chain | 32 | 5961.83 | |
| Collagen alpha-2(I) chain | 63 | 3161.86 | |
| Collagen alpha-1(XXII) chain | 44 | 3020.04 | |
| Collagen alpha-6(IV) chain | 24 | 2429.28 | |
| Collagen alpha-1(VII) chain | 56 | 1996.28 | |
| Collagen alpha-2(XI) chain | 52 | 1264.12 | |
| Collagen alpha-1(III) chain | 73 | 1181.17 | |
| Collagen alpha-1(V) chain | 34 | 1154.63 | |
| Collagen alpha-3(VI) chain | 72 | 869.6 |
| Parental Pathway | Enriched Pathway | Total Number of Proteins in the Pathway | No. of Assigned Proteins | FDR |
|---|---|---|---|---|
| Extracellular matrix organization | Extracellular matrix organization | 418 | 54 | 7.72 × 10−47 |
| Collagen chain trimerization | 44 | 40 | 6.68 × 10−78 | |
| Collagen biosynthesis and modifying enzymes | 67 | 41 | 3.81 × 10−67 | |
| Collagen formation | 90 | 43 | 3.34 × 10−64 | |
| Assembly of collagen fibrils and other multimeric structures | 61 | 33 | 3.09 × 10−51 | |
| Anchoring fibril formation | 15 | 9 | 2.18 × 10−14 | |
| Collagen degradation | 89 | 34 | 2.73 × 10−46 | |
| Degradation of the extracellular matrix | 188 | 39 | 1.43 × 10−41 | |
| Integrin cell surface interactions | 109 | 33 | 5.15 × 10−41 | |
| ECM proteoglycans | 90 | 24 | 5.12 × 10−28 | |
| Non-integrin membrane-ECM interactions | 73 | 18 | 2.42 × 10−20 | |
| Hemostasis | Hemostasis | 738 | 39 | 6.04 × 10−19 |
| Platelet degranulation | 128 | 25 | 1.35 × 10−25 | |
| Response to elevated platelet cytosolic Ca2+ | 133 | 25 | 3.44 × 10−25 | |
| Platelet activation signaling and aggregation | 295 | 27 | 7.54 × 10−19 | |
| Developmental Biology | NCAM signaling for neurite out-growth | 74 | 20 | 1.59 × 10−23 |
| NCAM1 interactions | 43 | 18 | 3.84 × 10−25 | |
| Metabolism of proteins | Regulation of Insulin-like Growth Factor IGF transport and uptake by Insulin-like Growth Factor Binding Proteins IGFBPs | 124 | 17 | 1.15 × 10−14 |
| Post-translational protein phosphorylation | 107 | 16 | 1.90 × 10−14 | |
| Signal Transduction | Signaling by Receptor Tyrosine Kinases | 555 | 29 | 7.95 × 10−14 |
| Signaling by PDGF | 58 | 20 | 7.84 × 10−26 | |
| MET activates PTK2 signaling | 30 | 11 | 1.50 × 10−14 | |
| MET promotes cell motility | 41 | 11 | 6.61 × 10−13 | |
| Immune System | Immune System | 2610 | 60 | 1.97 × 10−12 |
| Neutrophil degranulation | 495 | 26 | 1.97 × 10−12 |
| Name | # Peptides | Fold Change | |||
|---|---|---|---|---|---|
| Rough Surface: 5 Min vs. 2 Min | Smooth Surface: 5 Min vs. 2 Min | 2 Min Exposure: Rough vs. Smooth | 5 Min Exposure: Rough vs. Smooth | ||
| Intracellular, Membrane | |||||
| Probable E3 ubiquitin-protein ligase HECTD4 | 8 | 2.56 | 8.76 | 4.22 | 1.24 |
| Aldehyde dehydrogenase, mitochondrial | 10 | 0.37 | 8.33 | 26.44 | 1.16 |
| Glutathione S-transferase omega-1 | 8 | 0.39 | 6.53 | 2.18 | 0.13 |
| Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic | 9 | 0.89 | 5.05 | 9.33 | 1.65 |
| Heat shock protein HSP 90-beta | 10 | 0.27 | 2.57 | 4.49 | 0.48 |
| Hemoglobin subunit gamma-2 | 10 | 1.58 | 2.22 | 2.35 | 1.67 |
| Ankyrin-1 | 33 | 2.08 | 2.17 | 1.19 | 1.15 |
| Hemoglobin subunit beta | 25 | 0.93 | 0.54 | 0.95 | 1.65 |
| L-lactate dehydrogenase B chain | 12 | 0.99 | 0.34 | 1.36 | 3.91 |
| Myeloperoxidase | 19 | 0.40 | 0.29 | 2.03 | 2.83 |
| Rab GDP dissociation inhibitor beta | 8 | 0.07 | 0.06 | 4.07 | 4.63 |
| Secreted to blood | |||||
| Complement C5 | 11 | 1.36 | 5.60 | 9.18 | 2.23 |
| Plasminogen | 15 | 1.34 | 4.83 | 4.17 | 1.15 |
| Coagulation factor XIII A chain | 10 | 1.06 | 3.56 | 0.76 | 0.23 |
| Histidine-rich glycoprotein | 9 | 0.80 | 3.32 | 0.67 | 0.16 |
| Antithrombin-III | 15 | 0.94 | 2.16 | 1.48 | 0.65 |
| Leukocyte elastase inhibitor | 10 | 0.71 | 2.42 | 2.52 | 0.74 |
| ECM | |||||
| Mucin-19 | 10 | 1.86 | 9.41 | 11.43 | 2.26 |
| Filaggrin | 8 | 1.81 | 7.15 | 8.80 | 2.22 |
| Collagen alpha-1(III) chain | 73 | 0.45 | 6.01 | 6.89 | 0.52 |
| Collagen alpha-1(XXI) chain | 11 | 1.91 | 3.41 | 1.37 | 0.77 |
| Collagen alpha-1(XVIII) chain | 17 | 2.40 | 3.30 | 1.10 | 0.80 |
| Collagen alpha-2(IX) chain | 15 | 3.80 | 3.26 | 1.56 | 1.82 |
| Collagen alpha-1(XXII) chain | 44 | 0.97 | 2.87 | 2.92 | 0.99 |
| Collagen alpha-3(V) chain | 35 | 1.70 | 2.32 | 1.88 | 1.37 |
| Collagen alpha-1(XI) chain | 41 | 1.20 | 2.27 | 0.99 | 0.52 |
| Collagen alpha-6(VI) chain | 20 | 0.80 | 0.69 | 1.18 | 1.37 |
| Collagen alpha-4(IV) chain | 45 | 1.95 | 1.42 | 0.97 | 1.33 |
| Parental Pathway | Enriched Pathway | Total Number of Proteins in the Pathway | No. of Assigned Proteins | FDR |
|---|---|---|---|---|
| Extracellular matrix organization | Collagen formation | 90 | 9 | 1.14 × 10−3 |
| Collagen chain trimerization | 44 | 8 | 1.11 × 10−4 | |
| Collagen biosynthesis and modifying enzymes | 67 | 9 | 1.37 × 10−4 | |
| Assembly of collagen fibrils and other multimeric structures | 61 | 6 | 2.94 × 10−2 | |
| Hemostasis | Common pathway of fibrin clot formation | 22 | 4 | 2.94 × 10−2 |
| Developmental Biology | NCAM signaling for neurite out-growth | 74 | 7 | 1.47 × 10−2 |
| NCAM1 interactions | 43 | 5 | 3.56 × 10−2 |
| Name | # Peptides | Fold Change | |||
|---|---|---|---|---|---|
| 2 Min Exposure: Rough versus Smooth | 5 Min Exposure: Rough versus Smooth | Rough Surface: 5 Min vs. 2 Min | Smooth Surface: 5 Min vs. 2 Min | ||
| Intracellular, Membrane | |||||
| Nuclear receptor corepressor 2 | 8 | 3.31 | 2.40 | 0.99 | 1.36 |
| 14-3-3 protein epsilon | 10 | 2.99 | 3.45 | 1.04 | 0.90 |
| Endoplasmic reticulum chaperone BiP | 10 | 4.97 | 1.42 | 0.34 | 1.18 |
| Transitional endoplasmic reticulum ATPase | 13 | 3.87 | 1.82 | 0.93 | 1.96 |
| Eosinophil peroxidase | 9 | 8.63 | 1.85 | 0.41 | 1.91 |
| Four and a half LIM domains protein 1 | 8 | 5.03 | 4.08 | 0.63 | 0.78 |
| Rab GDP dissociation inhibitor beta | 8 | 4.07 | 4.63 | 0.07 | 0.06 |
| Filamin-A | 33 | 2.97 | 1.28 | 1.32 | 3.05 |
| Protein piccolo | 8 | 20.26 | 1.51 | 0.36 | 4.85 |
| Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic | 9 | 9.33 | 1.65 | 0.89 | 5.05 |
| Alcohol dehydrogenase 1B | 8 | 8.93 | 6.96 | 1.37 | 1.76 |
| Aldehyde dehydrogenase, mitochondrial | 10 | 26.44 | 1.16 | 0.37 | 8.33 |
| Protein bassoon | 9 | 2.82 | 1.44 | 0.58 | 1.14 |
| Tropomyosin beta chain | 17 | 3.20 | 6.02 | 0.37 | 0.20 |
| Myeloperoxidase | 19 | 2.03 | 2.83 | 0.40 | 0.29 |
| Malate dehydrogenase, cytoplasmic | 8 | 6.28 | 6.93 | 0.67 | 0.60 |
| Hemoglobin subunit delta | 12 | 2.84 | 2.58 | 0.86 | 0.95 |
| Histone H4 | 10 | 2.43 | 2.52 | 1.38 | 1.33 |
| Keratin, type II cytoskeletal 2 epidermal | 25 | 0.12 | 0.09 | 0.80 | 1.00 |
| Myosin-7 | 99 | 3.40 | 4.10 | 0.92 | 0.76 |
| Tubulin alpha-1B chain | 9 | 0.61 | 0.38 | 0.87 | 1.39 |
| Isocitrate dehydrogenase [NADP], mitochondrial | 10 | 14.95 | Only Rough | 0.44 | Only Rough |
| ADP/ATP translocase 1 | 8 | 5.88 | 22.48 | 0.72 | 0.19 |
| Protein 4.1 | 10 | 13.95 | 15.43 | 1.98 | 1.79 |
| Prelamin-A/C | 17 | 3.98 | 13.29 | 1.62 | 0.49 |
| ATP synthase subunit alpha, mitochondrial | 12 | 2.52 | 10.00 | 0.50 | 0.13 |
| Alpha-crystallin B chain | 8 | 21.87 | 7.80 | 1.33 | 3.73 |
| Myosin light chain 1/3, skeletal muscle isoform | 9 | 1.92 | 7.74 | 2.07 | 0.51 |
| Myosin light chain 3 | 9 | 2.13 | 6.33 | 0.52 | 0.17 |
| Coronin-1A | 8 | 10.30 | 6.11 | 0.27 | 0.46 |
| Triosephosphate isomerase | 12 | 6.40 | 6.06 | 0.84 | 0.89 |
| L-lactate dehydrogenase A chain | 9 | 3.04 | 4.56 | 1.51 | 1.01 |
| L-lactate dehydrogenase B chain | 12 | 1.36 | 3.91 | 0.99 | 0.34 |
| Glyceraldehyde-3-phosphate dehydrogenase | 12 | 0.95 | 3.79 | 0.89 | 0.22 |
| Carbonic anhydrase 2 | 16 | 1.38 | 3.75 | 0.60 | 0.22 |
| Transketolase | 14 | 3.14 | 3.35 | 1.42 | 1.32 |
| Vinculin | 8 | 3.24 | 3.22 | 1.27 | 1.28 |
| Erythrocyte membrane protein band 4.2 | 10 | 2.05 | 3.00 | 0.52 | 0.36 |
| Pyruvate kinase PKM | 10 | 2.06 | 2.85 | 1.57 | 1.13 |
| Alpha-enolase | 14 | 1.05 | 2.75 | 0.60 | 0.23 |
| Vimentin | 24 | 0.81 | 2.57 | 0.21 | 0.07 |
| Fructose-bisphosphate aldolase A | 13 | 2.17 | 2.50 | 0.99 | 0.86 |
| Hemoglobin subunit alpha | 25 | 1.11 | 2.46 | 0.87 | 0.40 |
| Phosphoglycerate kinase 1 | 9 | 1.97 | 2.36 | 1.40 | 1.16 |
| Peroxiredoxin-2 | 16 | 1.95 | 2.35 | 1.03 | 0.85 |
| Plectin | 11 | 0.05 | 2.11 | 2.05 | 0.05 |
| Actin, cytoplasmic 1 | 16 | 1.52 | 2.05 | 0.97 | 0.72 |
| Carbonic anhydrase 1 | 19 | 1.78 | 2.02 | 0.71 | 0.62 |
| Catalase | 25 | 1.66 | 1.98 | 0.98 | 0.82 |
| Hemoglobin subunit beta | 25 | 0.95 | 1.65 | 0.93 | 0.54 |
| Alpha-actinin-2 | 31 | 1.06 | 1.52 | 0.98 | 0.68 |
| Keratin, type I cytoskeletal 9 | 14 | 0.09 | 0.41 | 1.91 | 0.41 |
| Glutathione S-transferase omega-1 | 8 | 2.18 | 0.13 | 0.39 | 6.53 |
| Secreted to blood | |||||
| Fibronectin | 34 | 3.60 | 1.25 | 0.72 | 2.07 |
| Annexin A2 | 15 | 3.15 | 7.27 | 1.63 | 0.71 |
| Leukocyte elastase inhibitor | 10 | 2.52 | 0.74 | 0.71 | 2.42 |
| Plasminogen | 15 | 4.17 | 1.15 | 1.34 | 4.83 |
| Complement C5 | 11 | 9.18 | 2.23 | 1.36 | 5.60 |
| Afamin | 11 | 8.08 | 1.77 | 0.78 | 3.55 |
| Alpha-2-macroglobulin | 56 | 2.21 | 1.95 | 1.23 | 1.40 |
| Complement C3 | 85 | 2.59 | 2.39 | 1.19 | 1.28 |
| Complement factor B | 17 | 3.61 | 4.34 | 0.68 | 0.57 |
| Alpha-1-antitrypsin | 27 | 2.49 | 3.18 | 1.27 | 1.00 |
| Apolipoprotein A-I | 26 | 2.54 | 1.98 | 0.99 | 1.28 |
| Angiotensinogen | 9 | 2.73 | 1.81 | 1.38 | 2.08 |
| Complement C4-B | 43 | 4.36 | 4.06 | 1.02 | 1.10 |
| Antithrombin-III | 15 | 1.48 | 0.65 | 0.94 | 2.16 |
| Inter-alpha-trypsin inhibitor heavy chain H2 | 14 | 4.05 | 4.40 | 1.04 | 0.96 |
| Plasma kallikrein | 12 | 4.04 | 14.63 | 2.52 | 0.70 |
| Neutrophil elastase | 8 | 3.25 | 3.89 | 0.98 | 0.82 |
| Haptoglobin | 24 | 1.95 | 2.33 | 1.46 | 1.21 |
| Histidine-rich glycoprotein | 9 | 0.67 | 0.16 | 0.80 | 3.32 |
| Inter-alpha-trypsin inhibitor heavy chain H1 | 9 | 2.55 | 2.55 | 0.54 | 0.54 |
| Hemopexin | 17 | 0.74 | 1.57 | 1.13 | 0.54 |
| Coagulation factor XIII A chain | 10 | 0.76 | 0.23 | 1.06 | 3.56 |
| Apolipoprotein B-100 | 98 | 1.01 | 1.39 | 0.65 | 0.47 |
| Fibrinogen gamma chain | 26 | 0.73 | 0.49 | 0.86 | 1.28 |
| ECM | |||||
| Collagen alpha-1(II) chain | 67 | 1.76 | 1.34 | 1.21 | 1.59 |
| Collagen alpha-1(III) chain | 73 | 6.89 | 0.52 | 0.45 | 6.01 |
| Collagen alpha-3(IV) chain | 13 | 7.07 | 0.27 | 0.45 | 11.56 |
| Collagen alpha-1(XXVIII) chain | 17 | 2.75 | 1.99 | 0.81 | 1.12 |
| Filaggrin | 8 | 8.80 | 2.22 | 1.81 | 7.15 |
| Collagen alpha-6(IV) chain | 24 | 0.66 | 0.51 | 0.90 | 1.18 |
| Collagen alpha-1(XXII) chain | 44 | 2.92 | 0.99 | 0.97 | 2.87 |
| Collagen alpha-1(XV) chain | 9 | 3.71 | 2.62 | 0.72 | 1.03 |
| Decorin | 9 | 10.76 | 10.38 | 1.39 | 1.44 |
| Collagen alpha-1(XIII) chain | 27 | 2.26 | 2.04 | 0.75 | 0.83 |
| Collagen alpha-1(XIV) chain | 20 | 1.94 | 2.60 | 2.00 | 1.50 |
| Lumican | 8 | 1.47 | 13.21 | 2.18 | 0.24 |
| Collagen alpha-1(XI) chain | 41 | 0.99 | 0.52 | 1.20 | 2.27 |
| Parental Pathway | Enriched Pathway | Total Number of Proteins in the Pathway | After 2 Min Exposure | After 5 Min Exposure | ||
|---|---|---|---|---|---|---|
| FDR | No. of Assigned Proteins | FDR | No. of Assigned Proteins | |||
| Extracellular matrix organization | Extracellular matrix organization | 418 | 1.96 × 10−5 | 17 | 2.20 × 10−3 | 18 |
| Collagen chain trimerization | 44 | 2.40 × 10−6 | 8 | 3.34 × 10−2 | 4 | |
| Collagen biosynthesis and modifying enzymes | 67 | 1.87 × 10−5 | 8 | - | - | |
| Collagen formation | 90 | 1.87 × 10−5 | 9 | - | - | |
| Degradation of the extracellular matrix | 188 | 1.87 × 10−5 | 12 | 3.78 × 10−3 | 11 | |
| Non-integrin membrane-ECM interactions | 73 | 2.37 × 10−4 | 7 | - | - | |
| Collagen degradation | 89 | 7.86 × 10−4 | 7 | 2.15 × 10−2 | 6 | |
| ECM proteoglycans | 90 | 7.86 × 10−4 | 7 | - | - | |
| Assembly of collagen fibrils and other multimeric structures | 61 | 7.89 × 10−4 | 6 | - | - | |
| Integrin cell surface interactions | 109 | 9.73 × 10−3 | 6 | - | - | |
| Hemostasis | Hemostasis | 738 | 7.10 × 10−3 | 17 | 3.16 × 10−6 | 33 |
| Platelet degranulation | 128 | 1.65 × 10−4 | 9 | 1.16 × 10−6 | 14 | |
| Response to elevated platelet cytosolic Ca2+ | 133 | 1.99 × 10−4 | 9 | 1.72 × 10−6 | 14 | |
| Platelet activation signaling and aggregation | 295 | 7.10 × 10−3 | 10 | 1.50 × 10−4 | 17 | |
| Metabolism of proteins | Regulation of Insulin-like Growth Factor IGF transport and uptake by Insulin-like Growth Factor Binding Proteins IGFBPs | 124 | 4.27 × 10−3 | 7 | 7.15 × 10−4 | 10 |
| Signal Transduction | MET activates PTK2 signaling | 30 | 4.29 × 10−3 | 4 | - | - |
| MAPK family signaling cascades | 299 | - | - | 1.72 × 10−4 | 17 | |
| RAF/MAP kinase cascade | 248 | - | - | 1.88 × 10−5 | 17 | |
| MAPK1/MAPK3 signaling | 254 | - | - | 2.49 × 10−5 | 17 | |
| Immune System | Immune System | 2610 | 4.27 × 10−3 | 41 | 1.22 × 10−5 | 73 |
| Neutrophil degranulation | 495 | 3.63 × 10−5 | 18 | 9.16 × 10−11 | 34 | |
| Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation | 39 | 1.05 × 10−3 | 5 | 2.41 × 10−2 | 4 | |
| Interleukin-12 signaling | 56 | 4.29 × 10−3 | 5 | - | - | |
| Activation of C3 and C5 | 12 | 4.33 × 10−3 | 3 | - | - | |
| Innate Immune System | 1313 | 2.15 × 10−2 | 23 | 4.18 × 10−8 | 52 | |
| Signaling by Interleukins | 706 | 4.41 × 10−2 | 14 | 3.32 × 10−6 | 32 | |
| Cytokine Signaling in Immune system | 983 | - | - | 6.30 × 10−5 | 36 | |
| FLT3 Signaling | 275 | - | - | 6.37 × 10−5 | 17 | |
| Other interleukin signaling | 298 | - | - | 3.16 × 10−6 | 20 | |
| Muscle contraction | Muscle contraction | 216 | 4.29 × 10−3 | 9 | 3.16 × 10−6 | 17 |
| Smooth Muscle Contraction | 42 | 1.41 × 10−3 | 5 | 7.50 × 10−3 | 5 | |
| Striated Muscle Contraction | 36 | 7.10 × 10−3 | 4 | 6.01 × 10−11 | 12 | |
| Transport of small molecules | Erythrocytes take up oxygen and release carbon dioxide | 9 | - | - | 2.71 × 10−4 | 4 |
| Vesicle-mediated transport | Binding and Uptake of Ligands by Scavenger Receptors | 112 | 4.15 × 10−2 | 5 | 3.50 × 10−4 | 10 |
| Metabolism | Metabolism | 2262 | - | - | 6.39 × 10−8 | 73 |
| Metabolism of carbohydrates | 312 | - | - | 8.30 × 10−12 | 29 | |
| Glucose metabolism | 93 | - | - | 2.81 × 10−9 | 15 | |
| Gluconeogenesis | 34 | 4.26 × 10−2 | 3 | 1.16 × 10−8 | 10 | |
| Glycolysis | 73 | - | - | 1.33 × 10−7 | 12 | |
| Programmed Cell Death | Programmed Cell Death | 193 | - | - | 8.29 × 10−5 | 14 |
| Apoptosis | 186 | - | - | 6.23 × 10−5 | 14 | |
| Activation of BH3-only proteins | 32 | 5.02 × 10−3 | 4 | - | - | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jäger, M.; Latosinska, A.; Herten, M.; Busch, A.; Grupp, T.; Sowislok, A. The Implant Proteome—The Right Surgical Glue to Fix Titanium Implants In Situ. J. Funct. Biomater. 2022, 13, 44. https://doi.org/10.3390/jfb13020044
Jäger M, Latosinska A, Herten M, Busch A, Grupp T, Sowislok A. The Implant Proteome—The Right Surgical Glue to Fix Titanium Implants In Situ. Journal of Functional Biomaterials. 2022; 13(2):44. https://doi.org/10.3390/jfb13020044
Chicago/Turabian StyleJäger, Marcus, Agnieszka Latosinska, Monika Herten, André Busch, Thomas Grupp, and Andrea Sowislok. 2022. "The Implant Proteome—The Right Surgical Glue to Fix Titanium Implants In Situ" Journal of Functional Biomaterials 13, no. 2: 44. https://doi.org/10.3390/jfb13020044
APA StyleJäger, M., Latosinska, A., Herten, M., Busch, A., Grupp, T., & Sowislok, A. (2022). The Implant Proteome—The Right Surgical Glue to Fix Titanium Implants In Situ. Journal of Functional Biomaterials, 13(2), 44. https://doi.org/10.3390/jfb13020044