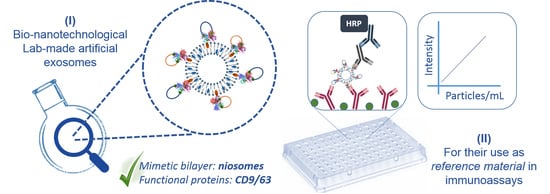
Selected Tetraspanins Functionalized Niosomes as Potential Standards for Exosome Immunoassays
Abstract
:1. Introduction
2. Materials and Methods
2.1. Niosomes Preparation and Size Measurement
2.2. Streptavidin Conjugation to Niosomes Surface
2.3. Tetraspanins (CD9/63) Large Extracellular Loops (LELs) Production
2.4. Vesicles Functionalization with Tetraspanins LELs Constructions
2.5. Immunoassays for Artificial EVs Detection
3. Results and Discussion
3.1. Streptavidin-Coated Niosomes Development as Generic Scaffold for Artificial EVs Production
3.2. Artificial EVs Production Using Nio_Str Functionalized with Tetraspanin LELs
3.3. Development of ELISA Assays Using Artificial Exosomes
3.3.1. Single Tetraspanin Functional Particles
3.3.2. Double Tetraspanin Functional Particles
3.4. Potential Commercial Use of Our Artificial Exosome Model
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yáñez-Mó, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 14, 27066. [Google Scholar] [CrossRef] [Green Version]
- Kalra, H.; Drummen, G.; Mathivanan, S. Focus on extracellular vesicles: Introducing the next small big thing. Int. J. Mol. Sci. 2016, 17, 170. [Google Scholar] [CrossRef] [Green Version]
- Azmi, A.S.; Bao, B.; Sarkar, F.H. Exosomes in cancer development, metastasis, and drug resistance: A comprehensive review. Cancer Metastasis Rev. 2013, 32, 623–642. [Google Scholar] [CrossRef] [Green Version]
- Corrado, C.; Raimondo, S.; Chiesi, A.; Ciccia, F.; De Leo, G.; Alessandro, R. Exosomes as intercellular signaling organelles involved in health and disease: Basic science and clinical applications. Int. J. Mol. Sci. 2013, 14, 5338–5366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Properzi, F.; Logozzi, M.; Fais, S. Exosomes: The future of biomarkers in medicine. Biomarkers Med. 2013, 7, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Cappello, F.; Logozzi, M.; Campanella, C.; Bavisotto, C.C.; Marcilla, A.; Properzi, F.; Fais, S. Exosome levels in human body fluids: A tumor marker by themselves? Eur. J. Pharm. Sci. 2017, 96, 93–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Meel, R.; Krawczyk-Durka, M.; van Solinge, W.W.; Schiffelers, R.M. Toward routine detection of extracellular vesicles in clinical samples. Int. J. Lab. Hematol. 2014, 36, 244–253. [Google Scholar] [CrossRef]
- Boriachek, K.; Islam, M.N.; Möller, A.; Salomon, C.; Nguyen, N.T.; Hossain, M.S.A.; Yamauchi, Y.; Shiddiky, M.J. Biological functions and current advances in isolation and detection strategies for exosome nanovesicles. Small 2018, 14, 1702153. [Google Scholar] [CrossRef]
- Peterson, M.F.; Otoc, N.; Sethi, J.K.; Gupta, A.; Antes, T.J. Integrated systems for exosome investigation. Methods 2015, 87, 31–45. [Google Scholar] [CrossRef]
- Van Der Pol, E.; Hoekstra, A.G.; Sturk, A.; Otto, C.; Van Leeuwen, T.G.; Nieuwland, R. Optical and non-optical methods for detection and characterization of microparticles and exosomes. J. Thromb. Haemost. 2010, 8, 2596–2607. [Google Scholar] [CrossRef]
- Rupert, D.L.; Lässer, C.; Eldh, M.; Block, S.; Zhdanov, V.P.; Lotvall, J.O.; Bally, M.; Höök, F. Determination of exosome concentration in solution using surface plasmon resonance spectroscopy. Anal. Chem. 2014, 86, 5929–5936. [Google Scholar] [CrossRef] [PubMed]
- Boriachek, K.; Islam, M.N.; Gopalan, V.; Lam, A.K.; Nguyen, N.T.; Shiddiky, M.J. Quantum dot-based sensitive detection of disease specific exosomes in serum. Analyst 2017, 142, 2211–2219. [Google Scholar] [CrossRef]
- Doldán, X.; Fagúndez, P.; Cayota, A.; Laíz, J.; Tosar, J.P. Electrochemical sandwich immunosensor for determination of exosomes based on surface marker-mediated signal amplification. Anal. Chem. 2016, 88, 10466–10473. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Rahimian, A.; Son, K.; Shin, D.S.; Patel, T.; Revzin, A. Development of an aptasensor for electrochemical detection of exosomes. Methods 2016, 97, 88–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreu, Z.; Yáñez-Mó, M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014, 16, 442. [Google Scholar] [CrossRef] [Green Version]
- Jang, S.C.; Kim, O.Y.; Yoon, C.M.; Choi, D.S.; Roh, T.Y.; Park, J.; Nilsson, J.; Lötvall, J.; Kim, Y.K.; Gho, Y.S. Bioinspired exosome-mimetic nanovesicles for targeted delivery of chemotherapeutics to malignant tumors. ACS Nano 2013, 7, 7698–7710. [Google Scholar] [CrossRef]
- Jeong, D.; Jo, W.; Yoon, J.; Kim, J.; Gianchandani, S.; Gho, Y.S.; Park, J. Nanovesicles engineered from ES cells for enhanced cell proliferation. Biomaterials 2014, 35, 9302–9310. [Google Scholar] [CrossRef]
- Hood, J.L.; Scott, M.J.; Wickline, S.A. Maximizing exosome colloidal stability following electroporation. Anal. Biochem. 2014, 448, 41–49. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Kim, J.; Jeong, M.; Lee, H.; Goh, U.; Kim, H.; Kim, B.; Park, J.H. Liposome-based engineering of cells to package hydrophobic compounds in membrane vesicles for tumor penetration. Nano Lett. 2015, 15, 2938–2944. [Google Scholar] [CrossRef]
- Lane, R.E.; Korbie, D.; Anderson, W.; Vaidyanathan, R.; Trau, M. Analysis of exosome purification methods using a model liposome system and tunable-resistive pulse sensing. Sci. Rep. 2015, 5, 7639. [Google Scholar] [CrossRef] [Green Version]
- Maas, S.L.; De Vrij, J.; Van Der Vlist, E.J.; Geragousian, B.; Van Bloois, L.; Mastrobattista, E.; Schiffelers, R.M.; Wauben, M.H.M.; Broekman, M.L.D.; Nolte, E.N. Possibilities and limitations of current technologies for quantification of biological extracellular vesicles and synthetic mimics. J. Control. Release 2015, 200, 87–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Manrique, P.; Gutiérrez, G.; Blanco-López, M.C. Fully artificial exosomes: Towards new theranostic biomaterials. Trends Biotechnol. 2018, 36, 10–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Manrique, P.; Matos, M.; Gutiérrez, G.; Pazos, C.; Blanco-López, M.C. Therapeutic biomaterials based on extracellular vesicles: Classification of bio-engineering and mimetic preparation routes. J. Extracell. Vesicles 2018, 7, 1422676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lozano-Andrés, E.; Libregts, S.F.; Toribio, V.; Royo, F.; Morales, S.; López-Martín, S.; Valés-Gómez, M.; Reyburn, H.T.; Falcón-Pérez, J.M.; Wauben, M.H.; et al. Tetraspanin-decorated extracellular vesicle-mimetics as a novel adaptable reference material. J. Extracell. Vesicles 2019, 8, 1573052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geeurickx, E.; Tulkens, J.; Dhondt, B.; Van Deun, J.; Lippens, L.; Vergauwen, G.; Heyrman, E.; De Sutter, D.; Gevaert, K.; Impens, F.; et al. The generation and use of recombinant extracellular vesicles as biological reference material. Nat. Commun. 2019, 10, 3288. [Google Scholar] [CrossRef] [Green Version]
- García-Manrique, P.; Matos, M.; Gutiérrez, G.; Estupiñán, O.R.; Blanco-López, M.C.; Pazos, C. Using factorial experimental design to prepare size-tuned nanovesicles. Ind. Eng. Chem. Res. 2016, 55, 9164–9175. [Google Scholar] [CrossRef] [Green Version]
- Hermanson, G.T. (Ed.) Introduction to Bioconjugation, and The Chemistry of Reactive Groups. In Bioconjugate Techniques, 2nd ed.; Academic Press: Londres, UK, 2008. [Google Scholar]
- Endoh, H.; Suzuki, Y.; Hashimoto, Y. Antibody coating of liposomes with 1-ethyl-3-(3-dimethyl-aminopropyl)carbodiimide and the effect on target specificity. J. Immunol. Methods 1981, 44, 79–85. [Google Scholar] [CrossRef]
- Tan, D.M.-Y.; Fu, J.-Y.; Fu-Shun Wong, F.-S.; Er, H.-M.; Chen, Y.-S.; Nesaretnam, K. Tumor regression and modulation of gene expression via tumor-targeted tocotrienol niosomes. Nanomedicine 2017, 12, 2487–2502. [Google Scholar] [CrossRef]
- Li, K.; Chang, S.; Wang, Z.; Zhao, X.; Chen, D. A novel micro-emulsion and micelle assembling method to prepare DEC205 monoclonal antibody coupled cationic nanoliposomes for simulating exosomes to target dendritic cells. Int. J. Pharm. 2015, 491, 105–112. [Google Scholar] [CrossRef]
- Sitar, S.; Kejžar, A.; Pahovnik, D.; Kogej, K.; Tušek-Žnidaric, M.; Lenassi, M.; Žagar, E. Size characterization and quantification of exosomes by Aymmetrical-Flow Field-Flow Fractionation. Anal. Chem. 2015, 87, 9225–9233. [Google Scholar] [CrossRef]
- Szatanek, R.; Baj-Krzyworzeka, M.; Zimoch, J.; Lekka, M.; Siedlar, M.; Baran, J. The methods of choice for Extracellular Vesicles (EVs) characterization. Int. J. Mol. Sci. 2017, 18, 1153. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Pertierra, E.; Oliveira-Rodríguez, M.; Rivas, M.; Oliva, P.; Villafani, J.; Navarro, A.; Blanco-López, M.C.; Cernuda-Morollón, E. Characterization of plasma-derived extracellular vesicles isolated by different methods: A comparison study. Bioengineering 2019, 6, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rong, Y.; Liu, W.; Wang, J.; Fan, J.; Luo, Y.; Li, L.; Kong, F.; Chen, J.; Tang, P.; Cai, W. Neural stem cell-derived small extracellular vesicles attenuate apoptosis and neuroinflammation after traumatic spinal cord injury by activating autophagy. Cell Death Dis. 2019, 10, 340. [Google Scholar] [CrossRef] [PubMed]
- Charoenviriyakul, C.; Takahashi, Y.; Nishikawa, M.; Takakura, Y. Preservation of exosomes at room temperature using lyophilization. Int. J. Pharm. 2018, 553, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Renart, J.; Behrens, M.M.; Fernández-Renart, M.; Martínez, J.L. Immunoassay; Christopoulos, T.K., Diamandis, E.P., Eds.; Academic Press Inc.: San Diego, CA, USA, 1996; pp. 537–554. ISBN 0080534503. [Google Scholar]
- Christopoulos, T.K.; Diamandis, E.P. Immunoassay; Christopoulos, T.K., Diamandis, E.P., Eds.; Academic Press Inc.: San Diego, CA, USA, 1996; pp. 227–236. ISBN 0080534503. [Google Scholar]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Florent Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA. 2016, 113, E968–E977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rupert, D.L.; Claudio, V.; Lässer, C.; Bally, M. Methods for the physical characterization and quantification of extracellular vesicles in biological samples. Biochim. Biophys. Acta Gen. Sub. 2017, 1861, 3164–3179. [Google Scholar] [CrossRef]
- Yoshioka, Y.; Konishi, Y.; Kosaka, N.; Katsuda, T.; Takashi, K.; Ochiya, T. Comparative marker analysis of extracellular vesicles in different human cancer types. J. Extracell. Vesicles 2013, 2, 20424. [Google Scholar] [CrossRef]






| Product | Manufacturer | Biomarkers | Assay Format | Standard Used for Calibration Plots |
|---|---|---|---|---|
| ExoELISA | SBI System Biosciences | CD9/CD63/CD81 for detection | Exosomes are immobilized directly into the well | Lyophilized Exosomes |
| ExoTest™ | HansaBioMed | CD9 for detection | Sandwich assay using CD9 for detection. Capture not specified by the manufacturer | Exosome lyophilized |
| ExoQuant | Centaur Genprice | CD9 for detection | Sandwich assay using pan-Exosome biomarkers (data not specify by the manufacturer | Lyophilized Exosomes |
| ExoEL-CD81A1 | BioVision | CD9 for detection | Sandwich assay using pan-Exosome biomarkers (data not specify by the manufacturer | Exosome lyophilized |
| PS Capture™ Exosome ELISA KIT | Fujufilm Wako Pure Chemical Corporation | CD63 for detection | Exosomes are captured by a phosphatidylserine binding protein immobilized in the wells | Lyophilized Exosomes |
| CD9/CD63 Exosome ELISA Kit | Cosmo BIO CO. Ltd. | CD63 for detection | Sandwich assay using CD9 for capture | CD9/63 Fusion protein |
| ExoAssay™ | CD Creative Diagnostics® | Not specified by the manufacturer | Sandwich assay using CD9 for capture | Lyophilized Exosomes |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Manrique, P.; Serrano-Pertierra, E.; Lozano-Andrés, E.; López-Martín, S.; Matos, M.; Gutiérrez, G.; Yáñez-Mó, M.; Blanco-López, M.C. Selected Tetraspanins Functionalized Niosomes as Potential Standards for Exosome Immunoassays. Nanomaterials 2020, 10, 971. https://doi.org/10.3390/nano10050971
García-Manrique P, Serrano-Pertierra E, Lozano-Andrés E, López-Martín S, Matos M, Gutiérrez G, Yáñez-Mó M, Blanco-López MC. Selected Tetraspanins Functionalized Niosomes as Potential Standards for Exosome Immunoassays. Nanomaterials. 2020; 10(5):971. https://doi.org/10.3390/nano10050971
Chicago/Turabian StyleGarcía-Manrique, Pablo, Esther Serrano-Pertierra, Estefanía Lozano-Andrés, Soraya López-Martín, María Matos, Gemma Gutiérrez, María Yáñez-Mó, and María Carmen Blanco-López. 2020. "Selected Tetraspanins Functionalized Niosomes as Potential Standards for Exosome Immunoassays" Nanomaterials 10, no. 5: 971. https://doi.org/10.3390/nano10050971
APA StyleGarcía-Manrique, P., Serrano-Pertierra, E., Lozano-Andrés, E., López-Martín, S., Matos, M., Gutiérrez, G., Yáñez-Mó, M., & Blanco-López, M. C. (2020). Selected Tetraspanins Functionalized Niosomes as Potential Standards for Exosome Immunoassays. Nanomaterials, 10(5), 971. https://doi.org/10.3390/nano10050971