Identification of a Profile of Neutrophil-Derived Granule Proteins in the Surface of Gold Nanoparticles after Their Interaction with Human Breast Cancer Sera
Abstract
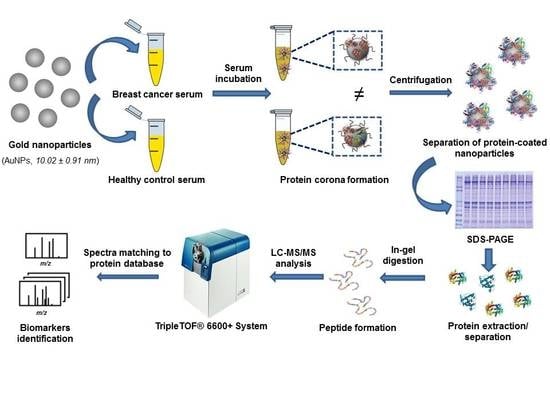
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Apparatus
2.3. Synthesis of Citrate-Gold Nanoparticles (AuNPs)
2.4. Sample Resources
2.5. Depletion, Reduction and Alkylation of Proteins Presented in Human Serum Samples (Healthy Controls and BC Patients)
2.6. Interaction of Proteins Presented in Human Serum Samples with the Surface of AuNPs: Formation of the Protein Corona (PC)
2.7. Separation of Serum Proteins Bound to the AuNPs Surface by 1-D gel Electrophoresis and Identification by Mass Spectrometry (LC-MS/MS)
2.8. Protein Functional Interaction Network Analysis and Protein Ontology Classification
3. Results and Discussion
3.1. Proteins Identified in the AuNP–protein Corona by Shotgun Proteomics Techniques
3.2. The Biological Role of the Proteins Identified in the AuNP–Protein Corona
Molecular Function and Pathway Analysis for Subtype Specific Breast Cancer
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, C.W.S.; Wu, M.; Cho, W.C.S.; To, K.K.W. Recent advances in the treatment of breast cancer. Front. Oncol. 2018, 8, 227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inic, Z.; Zegarac, M.; Inic, M.; Markovic, I.; Kozomara, Z.; Djurisic, I.; Inic, I.; Pupic, G.; Jancic, S. Difference between luminal A and luminal B subtypes according to Ki-67, tumor size, and progesterone receptor negativity providing prognostic information. Clin. Med. Insights Oncol. 2014, 8, 107–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Goldhirsch, A.; Winer, E.P.; Coates, A.S.; Gelber, R.D.; Piccart-Gebhart, M.; Thürlimann, B.; Senn, H.-J. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. 2013, 24, 2206–2223. [Google Scholar] [CrossRef]
- Coates, A.S.; Winer, E.P.; Goldhirsch, A.; Gelber, R.D.; Gnant, M.; Piccart-Gebhart, M.; Thürlimann, B.; Senn, H.-J. Tailoring therapies–improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann. Oncol. 2015, 26, 1533–1546. [Google Scholar] [CrossRef]
- Sotiriou, C.; Neo, S.Y.; McShane, L.M.; Korn, E.L.; Long, P.M.; Jazaeri, A.; Martiat, P.; Fox, S.B.; Harris, A.L.; Liu, E.T. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc. Natl. Acad. Sci. USA. 2003, 100, 10393–10398. [Google Scholar] [CrossRef] [Green Version]
- Nakshatri, H.; Qi, G.; You, J.; Kerry, B.; Schneider, B.; Zon, R.; Buck, C.; Regnier, F.; Wang, M. Intrinsic subtype-associated changes in the plasma proteome in breast cancer. Proteom. Clin. Appl. 2009, 3, 1305–1313. [Google Scholar] [CrossRef]
- Zhang, F.; Chen, J.Y. Breast cancer subtyping from plasma proteins. BMC Med. Genet. 2013, 6, S6. [Google Scholar] [CrossRef] [Green Version]
- Gajbhiye, A.; Dabhi, R.; Taunk, K.; Jagadeeshaprasad, M.G.; RoyChoudhury, S.; Mane, A.; Bayatigeri, S.; Chaudhury, K.; Santra, M.K.; Rapole, S. Multipronged quantitative proteomics reveals serum proteome alterations in breast cancer intrinsic subtypes. J. Proteome 2017, 163, 1–13. [Google Scholar] [CrossRef]
- Oda, M.; Makita, M.; Iwaya, K.; Akiyama, F.; Kohno, N.; Tsuchiya, B.; Iwase, T.; Matsubara, O. High levels of DJ 1 protein in nipple fluid of patients with breast cancer. Cancer Sci. 2012, 103, 1172–1176. [Google Scholar] [CrossRef] [PubMed]
- Somlo, G.; Lau, S.K.; Frankel, P.; Hsieh, H.B.; Liu, X.; Yang, L.; Krivacic, R.; Bruce, R.H. Multiple biomarker expression on circulating tumor cells in comparison to tumor tissues from primary and metastatic sites in patients with locally advanced/inflammatory, and stage IV breast cancer, using a novel detection technology. Breast Cancer Res. Treat. 2011, 128, 155–163. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.L.; Xiao, C.; Fu, G.; Wang, X.; Li, L. Identification of potential serum biomarkers for breast cancer using a functional proteomics technology. Biomark Res. 2017, 5, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- VanMeter, A.J.; Camerini, S.; Polci, M.L.; Tessitore, A.; Trivedi, N.; Heiby, M.; Kamal, Y.; Hansen, J.; Zhou, W. Serum low-molecular-weight protein fractionation for biomarker discovery. Methods Mol. Biol. 2012, 823, 237–249. [Google Scholar] [PubMed]
- Lee, P.Y.; Osman, J.; Low, T.Y.; Jamal, R. Plasma/serum proteomics: Depletion strategies for reducing high-abundance proteins for biomarker discovery. Bioanalysis 2019, 11, 1799–1812. [Google Scholar] [CrossRef]
- Chertov, O.; Simpson, J.T.; Biragyn, A.; Conrads, T.P.; Veenstra, T.D.; Fisher, R.J. Enrichment of low-molecular-weight proteins from biofluids for biomarker discovery. Expert Rev. Proteom. 2005, 2, 139–145. [Google Scholar] [CrossRef]
- Zhang, L.; Lu, H.; Yang, P. Recent developments of nanoparticle-based enrichment methods for mass spectrometric analysis in proteomic. Sci. China Chem. 2010, 53, 695–703. [Google Scholar] [CrossRef]
- Docter, D.; Westmeier, D.; Markiewicz, M.; Stolte, S.; Knauer, S.K.; Stauber, R.H. The nanoparticle biomolecule corona: Lessons learned—challenge accepted? Chem. Soc. Rev. 2015, 44, 6094–6121. [Google Scholar] [CrossRef] [Green Version]
- Lundqvist, M.; Stigler, J.; Elia, G.; Lynch, I.; Cedervall, T.; Dawson, K.A. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl. Acad. Sci. USA 2008, 105, 14265–14270. [Google Scholar] [CrossRef] [Green Version]
- Mahmoudi, M.; Lynch, I.; Ejtehadi, M.R.; Monopoli, M.P.; Bombelli, F.B.; Laurent, S. Protein-nanoparticle interactions: Opportunities and challenges. Chem. Rev. 2011, 111, 5610–5637. [Google Scholar] [CrossRef] [PubMed]
- Monopoli, M.P.; Walczyk, D.; Campbell, A.; Elia, G.; Lynch, I.; Baldelli Bombelli, F.; Dawson, K.A. Physical-chemical aspects of protein corona: Relevance to in vitroand in vivo biological impacts of nanoparticles. J. Am. Chem. Soc. 2011, 133, 2525–2534. [Google Scholar] [CrossRef] [PubMed]
- Laurent, S.; Burtea, C.; Thirifays, C.; Rezaee, F.; Mahmoudi, M. Significance of cell “observer” and protein source in nanobiosciences. J. Colloid Interface Sci. 2013, 392, 431–445. [Google Scholar] [CrossRef] [PubMed]
- Pozzi, D.; Caracciolo, G.; Digiacomo, L.; Colapicchioni, V.; Palchetti, S.; Capriotti, A.L.; Cavaliere, C.; Zenezini Chiozzi, R.; Puglisi, A.; Laganà, A. The biomolecular corona of nanoparticles in circulating biological media. Nanoscale 2015, 7, 13958–13966. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, M.; Abdelmonem, A.M.; Behzadi, S.; Clement, J.H.; Dutz, S.; Ejtehadi, M.R.; Hartmann, R.; Kantner, K.; Linne, U.; Maffre, P.; et al. Temperature: The “ignored” factor at the NanoBio interface. ACS Nano. 2013, 7, 6555–6562. [Google Scholar] [CrossRef]
- Tenzer, S.; Docter, D.; Kuharev, J.; Musyanovych, A.; Fetz, V.; Hecht, R.; Schlenk, F.; Fischer, D.; Kiouptsi, K.; Reinhardt, C.; et al. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotechnol. 2013, 8, 772–781. [Google Scholar] [CrossRef]
- Núñez, C.; Chantada-Vázquez, M.P.; Vázquez-Estévez, S. Inorganic nanoparticles in diagnosis and treatment of breast cancer. J. Biol. Inorg. Chem. 2018, 23, 331–345. [Google Scholar] [CrossRef]
- Núñez, C.; Chantada-Vázquez, M.P.; Bravo, S.B.; Vázquez-Estévez, S. Novel functionalized nanomaterials for the effective enrichment of proteins and peptides with post-translational modifications. J. Proteom. 2018, 181, 170–189. [Google Scholar] [CrossRef]
- Chantada-Vázquez, M.P.; Castro, A.; Bravo, S.B.; Vázquez-Estévez, S.; Acea-Nebril, B.; Núñez, C. Proteomic analysis of the bio-corona formed on the surface of (Au, Ag, Pt)-nanoparticles in human serum. Colloids Surf. B Biointerfaces 2019, 177, 141–148. [Google Scholar]
- Núñez, C. Blood-based protein biomarkers in breast cancer. Clin. Chim. Acta 2019, 490, 113–127. [Google Scholar] [CrossRef]
- García Vence, M.; Chantada-Vázquez, M.P.; Vázquez-Estévez, S.; Cameselle-Teijeiro, J.M.; Bravo, S.B.; Núñez, C. Potential clinical applications of the personalized, disease-specific protein corona on nanoparticles. Clin. Chim. Acta 2020, 501, 102–111. [Google Scholar]
- Chantada-Vázquez, M.P.; Castro, A.; García Vence, M.; Vázquez-Estévez, S.; Acea-Nebril, B.; Calatayud, D.G.; Jardiel, T.; Bravo, S.B.; Núñez, C. Proteomic investigation on bio-corona of Au, Ag and Fe nanoparticles for the discovery of triple negative breast cancer serum protein biomarkers. J. Proteom. 2020, 212, 103581. [Google Scholar]
- Couto, C.; Vitorino, R.; Daniel-da-Silva, A.L. Gold nanoparticles and bioconjugation: A pathway for proteomic applications. Crit. Rev. Biotechnol. 2017, 37, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Cortés, R.; Oliveira, E.; Núñez, C.; Lodeiro, C.; Páez de la Cadena, M.; Fdez-Riverola, F.; López-Fernández, H.; Reboiro-Jato, M.; Glez-Peña, D.; Capelo, J.L.; et al. Fast human serum profiling through chemical depletion coupled to gold-nanoparticle-assisted protein separation. Talanta 2012, 100, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Warder, S.E.; Tucker, L.A.; Strelitzer, T.J.; McKeegan, E.M.; Meuth, J.L.; Jung, P.M.; Saraf, A.; Singh, B.; Lai-Zhang, J.; Gagne, G.; et al. Reducing agent-mediated precipitation of high-abundance plasma proteins. Anal. Biochem. 2009, 387, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Fernández, C.; Santos, H.M.; Ruíz-Romero, C.; Blanco, F.J.; Capelo-Martínez, J.L. A comparison of depletion versus equalization for reducing high-abundance proteins in human serum. Electrophoresis 2011, 32, 2966–2974. [Google Scholar]
- Candiano, G.; Bruschi, M.; Musante, L.; Santucci, L.; Ghiggeri, G.M.; Carnemolla, B.; Orecchia, P.; Zardi, L.; Righetti, P.G. Blue silver: A very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 2004, 25, 1327–1333. [Google Scholar] [CrossRef]
- Oliveira, E.; Araújo, J.E.; Gómez-Meire, S.; Lodeiro, C.; Pérez-Melón, C.; Iglesias-Lamas, E.; Otero-Glez, A.; Capelo, J.L.; Santos, H.M. Proteomics analysis of the peritoneal dialysate effluent reveals the presence of calcium-regulation proteins and acute inflammatory response. Clin. Proteom. 2014, 11, 17. [Google Scholar] [CrossRef] [Green Version]
- Shilov, I.V.; Seymour, S.L.; Patel, A.A.; Loboda, A.; Tang, W.H.; Keating, S.P.; Hunter, C.L.; Nuwaysir, L.M.; Schaeffer, D.A. The paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell Proteom. 2007, 6, 1638–1655. [Google Scholar] [CrossRef] [Green Version]
- Tang, W.H.; Shilov, I.V.; Seymour, S.L. Nonlinear fitting method for determining local false discovery rates from decoy database searches. J. Proteome Res. 2008, 7, 3661–3667. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Dobrovolskaia, M.A.; Patri, A.K.; Zheng, J.; Clogston, J.D.; Ayub, N.; Aggarwal, P.; Neun, B.W.; Hall, J.B.; McNeil, S.E. Interaction of colloidal gold nanoparticles with human blood: Effects on particle size and analysis of plasma protein binding profiles. Nanomedicine 2009, 5, 106–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Álvarez, R.; Hadjidemetriou, M.; Sánchez-Iglesias, A.; Liz-Marzán, L.M.; Kostarelos, K. In vivo formation of protein corona on gold nanoparticles. The effect of size and shape. Nanoscale 2018, 10, 1256–1264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fry, S.A.; Sinclair, J.; Timms, J.F.; Leathem, A.J.; Dwek, M.V. A targeted glycoproteomic approach identifies cadherin-5 as a novel biomarker of metastatic breast cancer. Cancer Lett. 2013, 328, 335–344. [Google Scholar] [CrossRef]
- Fry, S.A.; Robertson, C.E.; Swann, R.; Dwek, M.V. Cadherin-5: A biomarker for metastatic breast cancer with optimum efficacy in oestrogen receptor-positive breast cancers with vascular invasion. Br. J. Cancer 2016, 114, 1019–1026. [Google Scholar] [CrossRef] [PubMed]
- Cassatella, M.A.; Östberg, N.K.; Tamassia, N.; Soehnlein, O. Biological Roles of Neutrophil-Derived Granule Proteins and Cytokines. Trends Immunol. 2019, 40, 648–664. [Google Scholar] [CrossRef]
- Mollinedo, F. Neutrophil Degranulation, Plasticity, and Cancer Metastasis. Trends Immunol. 2019, 40, 228–242. [Google Scholar] [CrossRef]
- Borregaard, N.; Sørensen, O.E.; Theilgaard-Mönch, K. Neutrophil granules: A library of innate immunity proteins. Trends Immunol. 2007, 28, 340–345. [Google Scholar] [CrossRef]
- Mollinedo, F.; Pulido, R.; Lacal, P.M.; Sánchez-Madrid, F. Mobilization of gelatinase-rich granules as a regulatory mechanism of early functional responses in human neutrophils. Scand. J. Immunol. 1991, 34, 33–43. [Google Scholar] [CrossRef]
- Deryugina, E.I.; Zajac, E.; Juncker-Jensen, A.; Kupriyanova, T.A.; Welter, L.; Quigley, J.P. Tissue-infiltrating neutrophils constitute the major in vivo source of angiogenesis-inducing MMP-9 in the tumor microenvironment. Neoplasia 2014, 16, 771–788. [Google Scholar] [CrossRef] [Green Version]
- Bundred, N.J.; Dover, M.S.; Aluwihare, N.; Faragher, E.B.; Morrison, J.M. Smoking and periductal mastitis. BMJ. 1993, 307, 772–773. [Google Scholar] [CrossRef] [Green Version]
- Josephy, P.D. The role of peroxidase-catalyzed activation of aromatic amines in breast cancer. Mutagenesis 1996, 11, 3–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samoszuk, M.K.; Nguyen, V.; Gluzman, I.; Pham, J.H. Occult deposition of eosinophil peroxidase in a subset of human breast carcinomas. Am. J. Pathol. 1996, 148, 701–706. [Google Scholar] [PubMed]
- Panagopoulos, V.; Leach, D.A.; Zinonos, I.; Ponomarev, V.; Licari, G.; Liapis, V.; Ingman, W.V.; Anderson, P.; DeNichilo, M.O.; Evdokiou, A. Inflammatory peroxidases promote breast cancer progression in mice via regulation of the tumour microenvironment. Int. J. Oncol. 2017, 50, 1191–1200. [Google Scholar] [CrossRef] [Green Version]
- Wirthmueller, U.; Dewald, B.; Thelen, M.; Schäfer, M.K.; Stover, C.; Whaley, K.; North, J.; Eggleton, P.; Reid, K.B.; Schwaeble, W.J. Properdin, a positive regulator of complement activation, is released from secondary granules of stimulated peripheral blood neutrophils. J. Immunol. 1997, 158, 4444–4451. [Google Scholar] [PubMed]
- Wrighl, D.G. The neutrophil as a secretory organ of host defense. In Advances in Host Defense Mechanisms; vol. 1, Phagocytic cells; Gallin, J.L., Fauci, A.S., Eds.; Raven Press: New York, NY, USA, 1982; pp. 75–110. [Google Scholar]
- Nitto, T.; Onodera, K. Linkage between coenzyme A metabolism and inflammation: Roles of pantetheinase. J. Pharmacol. Sci. 2013, 123, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bjerrum, O.W.; Bjerrum, O.J.; Borregaard, N. Beta 2-microglobulin in neutrophils: An intragranular protein. J. Immunol. 1987, 138, 3913–3917. [Google Scholar]
- Gebhardt, C.; Németh, J.; Angel, P. Hess, S100A8 and S100A9 in inflammation and cancer. J. Biochem. Pharmacol. 2006, 72, 1622–1631. [Google Scholar] [CrossRef]
- Klein, T.; Levin, I.; Niska, A.; Koren, R.; Gal, R.; Schachter, J.; Kfir, B.; Narinski, R.; Warchaizer, S.; Klein, B. Correlation between tumour and serum beta 2m expression in patients with breast cancer. Eur. J. Immunogenet. 1996, 23, 417–423. [Google Scholar] [CrossRef]
- Madjd, Z.; Spendlove, I.; Pinder, S.E.; Ellis, I.O.; Durrant, L.G. Total loss of MHC class I is an independent indicator of good prognosis in breast cancer. Int. J. Cancer 2005, 117, 248–255. [Google Scholar] [CrossRef] [Green Version]
- Scheffer, G.L.; de Jong, M.C.; Monks, A.; Flens, M.J.; Hose, C.D.; Izquierdo, M.A.; Shoemaker, R.H.; Scheper, R.J. Increased expression of beta 2-microglobulin in multidrug-resistant tumour cells. Br. J. Cancer. 2002, 86, 1943–1950. [Google Scholar] [CrossRef] [Green Version]
- Moon, A.; Yong, H.Y.; Song, J.I.; Cukovic, D.; Salagrama, S.; Kaplan, D.; Putt, D.; Kim, H.; Dombkowski, A.; Kim, H.R. Global gene expression profiling unveils S100A8/A9 as candidate markers in H-ras-mediated human breast epithelial cell invasion. Mol. Cancer Res. 2008, 6, 1544–1553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, C.; Li, H.; Zhang, B.; Liu, Y.; Lu, G.; Lu, S.; Sun, L.; Qi, Y.; Li, X.; Chen, W. RAGE-binding S100A8/A9 promotes the migration and invasion of human breast cancer cells through actin polymerization and epithelial-mesenchymal transition. Breast Cancer Res. Treat. 2013, 142, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, H.; Petersson, S.; Enerback, C. Cluster analysis of S100 gene expression and genes correlating to psoriasin (S100A7) expression at different stages of breast cancer development. Int. J. Oncol. 2005, 27, 1473–1481. [Google Scholar] [PubMed]
- Rhee, D.K.; Park, S.H.; Jang, Y.K. Molecular signatures associated with transformation and progression to breast cancer in the isogenic MCF10 model. Genomics 2008, 92, 419–428. [Google Scholar] [CrossRef] [Green Version]
- Bao, Y.; Wang, A.; Mo, J. S100A8/A9 is associated with estrogen receptor loss in breast cancer. Oncol. Lett. 2016, 11, 1936–1942. [Google Scholar] [CrossRef] [Green Version]
- Naleskina, L.A.; Lukianova, N.Y.; Sobchenko, S.O.; Storchai, D.M.; Chekhun, V.F. Lactoferrin expression in breast cancer in relation to biologic properties of tumors and clinical features of disease. Exp. Oncol. 2016, 38, 181–186. [Google Scholar] [CrossRef]
- Hoedt, E.; Chaoui, K.; Huvent, I.; Mariller, C.; Monsarrat, B.; Burlet-Schiltz, O.; Pierce, A. SILAC-based proteomic profiling of the human MDA-MB-231 metastatic breast cancer cell line in response to the two antitumoral lactoferrin isoforms: The secreted lactoferrin and the intracellular delta-lactoferrin. PLoS ONE 2014, 9, e104563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahanta, S.; Paul, S.; Srivastava, A.; Pastor, A.; Kundu, B.; Chaudhuri, T.K. Stable self-assembled nanostructured hen egg white lysozyme exhibits strong anti-proliferative activity against breast cancer cells. Colloids Surf. B Biointerfaces 2015, 130, 237–245. [Google Scholar] [CrossRef]
- Udby, L.; Cowland, J.B.; Johnsen, A.H.; Sorensen, O.E.; Borregaard, N.; Kjeldsen, L. An ELISA for SGP28/ CRISP-3, a cysteine-rich secretory protein in human neutrophils, plasma, and exocrine secretions. J. Immunol. Methods 2002, 263, 43–55. [Google Scholar] [CrossRef]
- Wang, Y.; Sheng, N.; Xie, Y.; Chen, S.; Lu, J.; Zhang, Z.; Shan, Q.; Wu, D.; Zheng, G.; Li, M.; et al. Low expression of CRISP3 predicts a favorable prognosis in patients with mammary carcinoma. J. Cell Physiol. 2019, 234, 13629–13638. [Google Scholar] [CrossRef]
- Harris, J.P.; Caleb, M.H.; South, M.A. Secretory component in human mammary carcinoma. Cancer Res. 1975, 35, 1861–1864. [Google Scholar] [PubMed]
- Nyberg, P.; Ylipalosaari, M.; Sorsa, T.; Salo, T. Trypsins and their role in carcinoma growth. Exp. Cell Res. 2006, 312, 1219–1228. [Google Scholar] [CrossRef]
- Kehlen, A.; Lendeckel, U.; Dralle, H.; Langner, J.; Hoang-Vu, C. Biological significance of aminopeptidase N/CD13 in thyroid carcinomas. Cancer Res. 2003, 63, 8500–8506. [Google Scholar] [PubMed]
- Qian, L.; Gao, X.; Huang, H.; Lu, S.; Cai, Y.; Hua, Y.; Liu, Y.; Zhang, J. PRSS3 is a prognostic marker in invasive ductal carcinoma of the breast. Oncotarget 2017, 8, 21444–21453. [Google Scholar] [CrossRef] [Green Version]
- Dixon, J.; Kaklamanis, L.; Turley, H.; Hickson, I.D.; Leek, R.D.; Harris, A.L.; Gatter, K.C. Expression of aminopeptidase-N (CD 13) in normal tissues and malignant neoplasms of epithelial and lymphoid origin. J. Clin. Pathol. 1994, 47, 43–47. [Google Scholar] [CrossRef] [Green Version]
- Ranogajec, I.; Jakić-Razumović, J.; Puzović, V.; Gabrilovac, J. Prognostic value of matrix metalloproteinase-2 (MMP-2), matrix metalloproteinase-9 (MMP-9) and aminopeptidase N/CD13 in breast cancer patients. Med. Oncol. 2012, 29, 561–569. [Google Scholar] [CrossRef]
- Bai, Z.; Ye, Y.; Liang, B.; Feng, X.; Zhang, H.; Zhang, Y.; Peng, J.; Shen, D.; Cui, Z.; Zhang, Z.; et al. Proteomics-based identification of a group of apoptosis-related proteins and biomarkers in gastric cancer. Int. J. Oncol. 2011, 38, 375–383. [Google Scholar]
- Lee, D.H.; Chung, K.; Song, J.A.; Kim, T.H.; Kang, H.; Huh, J.H.; Jung, S.G.; Ko, J.J.; An, H.J. Proteomic identification of paclitaxel-resistance associated hnRNP A2 and GDI 2 proteins in human ovarian cancer cells. J. Proteome Res. 2010, 9, 5668–5676. [Google Scholar] [CrossRef]
- Zhang, P.; Garnett, J.; Creighton, C.J.; Al Sannaa, G.A.; Igram, D.R.; Lazar, A.; Liu, X.; Liu, C.; Pollock, R.E. EZH2-miR-30d-KPNB1 pathway regulates malignant peripheral nerve sheath tumour cell survival and tumourigenesis. J. Pathol. 2014, 232, 308–318. [Google Scholar] [CrossRef] [Green Version]
- Hendrix, A.; Braems, G.; Bracke, M.; Seabra, M.; Gahl, W.; De Wever, O.; Westbroek, W. The secretory small GTPase Rab27B as a marker for breast cancer progression. Oncotarget 2010, 1, 304–308. [Google Scholar] [CrossRef]
- Yang, P.S.; Yin, P.H.; Tseng, L.M.; Yang, C.H.; Hsu, C.Y.; Lee, M.Y.; Horng, C.F.; Chi, C.W. Rab5A is associated with axillary lymph node metastasis in breast cancer patients. Cancer Sci. 2011, 102, 2172–2178. [Google Scholar] [CrossRef]
- van der Watt, P.J.; Stowell, C.L.; Leaner, V.D. The nuclear import receptor Kpnβ1 and its potential as an anticancer therapeutic target. Crit. Rev. Eukaryot. Gene Expr. 2013, 23, 1–10. [Google Scholar] [CrossRef]
- de la Cruz-Merino, L.; Barco-Sánchez, A.; Henao Carrasco, F.; Nogales Fernández, E.; Vallejo Benítez, A.; Brugal Molina, J.; Martínez Peinado, A.; Grueso López, A.; Ruiz Borrego, M.; Codes Manuel de Villena, M.; et al. New insights into the role of the immune microenvironment in breast carcinoma. Clin. Dev. Immunol. 2013, 2013, 785317. [Google Scholar] [CrossRef]
- Oda, K.; Matsuoka, Y.; Funahashi, A.; Kitano, H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol. Syst. Biol. 2005, 1, 2005.0010. [Google Scholar] [CrossRef] [Green Version]
- Sobhani, N.; Fan, C.; Flores-Villanueva, P.O.; Generali, D.; Li, Y. The fibroblast growth factor receptors in breast cancer: From oncogenesis to better treatments. Int. J. Mol. Sci. 2020, 21, 2011. [Google Scholar] [CrossRef] [Green Version]






| Characteristics/Patients | Number | |
|---|---|---|
| Age (years) | <40 | 4 |
| 40–59 | 21 | |
| 60–80 | 16 | |
| >80 | 1 | |
| Tumor size (cm) | <2 | 25 |
| 2–5 | 14 | |
| >5 | 3 | |
| Histological types | In situ ductal carcinoma | 2 |
| Invasive ductal carcinoma | 36 | |
| In situ lobular carcinoma | 1 | |
| Invasive lobular carcinoma | 3 | |
| Receptor status | Luminal A | 11 |
| Luminal B-HER2 negative | 10 | |
| Luminal B-HER2 positive | 7 | |
| HER2 positive | 6 | |
| Triple negative | 8 | |
| Clinical stage | I | 15 |
| II | 20 | |
| III | 7 | |
| Nodal status | N0 | 25 |
| N1 | 17 | |
| LA (n = 29) | LB− (n = 41) | LB+ (n = 22) | HER2+ (n = 15) | TNBC (n = 23) | ||
|---|---|---|---|---|---|---|
| LYZ | CRISP3 | CTAGE9 | GAPDH | IGLV3-9 | PNMA6A | FAM110A |
| CRP | NEFH | KRT6C | GDI2 | TNS3 | CFAP100 | PLD5 |
| FILIP1L | SORBS1 | IGHV7-4-1 | MOAP1 | ANPEP | TOP1 | COG4 |
| BST1 | DCD | IGHV1-69D | AKAP9 | IFT140 | PRSS3P2 | PTPRD |
| TFAP2E | KRT15 | LRP2BP | FSIP2 | TPR | OTOG | ZNF404 |
| EXOC7 | ALCAM | IGHV4-30-2 | IGHV3-73 | STXBP5L | ADAP2 | SMC6 |
| EIF3C | ADIPOQ | BRPF3 | FGD6 | SFTPB | SLC9A1 | TRIM7 |
| N/A | MPO | BLVRB | KRT31 | SHC3 | ZNF426 | IGHV4-39 |
| TTN | PHLDA1 | HBD | KDM3B | TTC7A | MARK4 | SUPT20H |
| WEE1 | ABCB5 | PLCH1 | DES | MMP15 | TSBP1 | KMT2E |
| ACTBL2 | LDHAL6A | AK6 | CCDC28A | TBC1D1 | SETD1A | PPCS |
| TACC2 | KIF5B | IGHV3-53 | EPHB3 | EPAS1 | MMP12 | MYO15A |
| ANXA4 | PGLS | AK1 | CPD | GTPBP8 | WFDC3 | GRXCR2 |
| NSUN6 | N4BP1 | HBG1 | ELP3 | IGHV3-20 | SPATA9 | SNX25 |
| PNMA8C | PTPRG | CA3 | CPEB4 | ZGRF1 | ZNF622 | |
| SELENBP1 | FFAR4 | CCDC168 | ASB7 | |||
| RPS6KA3 | F7 | DNAH3 | KIF5A | |||
| MYH15 | MLLT1 | PRDM5 | IGHV3-21 | |||
| GSTO1 | PRDX2 | S100A9 | FGB | |||
| CPNE7 | IGKV1-8 | HHIPL2 | MCF2L2 | |||
| KPNB1 | S100A8 | CCDC106 | ||||
| LCP1 | NRXN3 | |||||
| IRF7 | ||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chantada-Vázquez, M.d.P.; García-Vence, M.; Vázquez-Estévez, S.; Bravo, S.B.; Núñez, C. Identification of a Profile of Neutrophil-Derived Granule Proteins in the Surface of Gold Nanoparticles after Their Interaction with Human Breast Cancer Sera. Nanomaterials 2020, 10, 1223. https://doi.org/10.3390/nano10061223
Chantada-Vázquez MdP, García-Vence M, Vázquez-Estévez S, Bravo SB, Núñez C. Identification of a Profile of Neutrophil-Derived Granule Proteins in the Surface of Gold Nanoparticles after Their Interaction with Human Breast Cancer Sera. Nanomaterials. 2020; 10(6):1223. https://doi.org/10.3390/nano10061223
Chicago/Turabian StyleChantada-Vázquez, María del Pilar, María García-Vence, Sergio Vázquez-Estévez, Susana B. Bravo, and Cristina Núñez. 2020. "Identification of a Profile of Neutrophil-Derived Granule Proteins in the Surface of Gold Nanoparticles after Their Interaction with Human Breast Cancer Sera" Nanomaterials 10, no. 6: 1223. https://doi.org/10.3390/nano10061223
APA StyleChantada-Vázquez, M. d. P., García-Vence, M., Vázquez-Estévez, S., Bravo, S. B., & Núñez, C. (2020). Identification of a Profile of Neutrophil-Derived Granule Proteins in the Surface of Gold Nanoparticles after Their Interaction with Human Breast Cancer Sera. Nanomaterials, 10(6), 1223. https://doi.org/10.3390/nano10061223