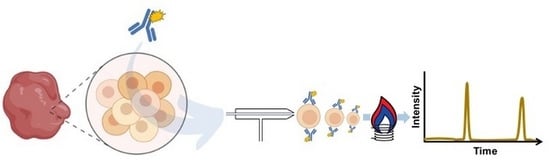
Capabilities of Single Cell ICP-MS for the Analysis of Cell Suspensions from Solid Tissues
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.2. Cell Cultures
2.3. Animal Models
2.4. Treatment with Accutase
2.5. Single Cell ICP-MS and Additional Instrumentation
2.6. Cell Fixation and Labelling with TfR1 Antibody
2.7. Data Treatment
3. Results and Discussion
3.1. Optimization of the Disaggregation Treatment with AccutaseTM
3.2. Analysis of Constitutive Elements by Single Cell ICP-MS Analysis
3.3. Analysis of TfR1 in the Cell Surface by Single Cell ICP-MS
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Altschuler, S.J.; Wu, L.F. Cellular Heterogeneity: Do Differences Make a Difference? Cell 2010, 141, 559–563. [Google Scholar] [CrossRef] [Green Version]
- Shapiro, E.; Biezuner, T.; Linnarsson, S. Single-Cell Sequencing-Based Technologies Will Revolutionize Whole-Organism Science. Nat. Rev. Genet. 2013, 14, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Corte-Rodríguez, M.; Álvarez-Fernández, R.; García-Cancela, P.; Montes-Bayón, M.; Bettmer, J. Single Cell ICP-MS Using on Line Sample Introduction Systems: Current Developments and Remaining Challenges. TrAC—Trends Anal. Chem. 2020, 132, 116042. [Google Scholar] [CrossRef]
- Wang, H.; Wang, B.; Wang, M.; Zheng, L.; Chen, H.; Chai, Z.; Zhao, Y.; Feng, W. Time-Resolved ICP-MS Analysis of Mineral Element Contents and Distribution Patterns in Single Cells. Analyst 2015, 140, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Theiner, S.; Loehr, K.; Koellensperger, G.; Mueller, L.; Jakubowski, N. Single-Cell Analysis by Use of ICP-MS. J. Anal. At. Spectrom. 2020, 35, 1784–1813. [Google Scholar] [CrossRef]
- Löhr, K.; Borovinskaya, O.; Tourniaire, G.; Panne, U.; Jakubowski, N. Arraying of Single Cells for Quantitative High Throughput Laser Ablation ICP-TOF-MS. Anal. Chem. 2019, 91, 11520–11528. [Google Scholar] [CrossRef]
- Van Malderen, S.J.M.; Van Acker, T.; Vanhaecke, F. Sub-Micrometer Nanosecond LA-ICP-MS Imaging at Pixel Acquisition Rates above 250 Hz via a Low-Dispersion Setup. Anal. Chem. 2020, 92, 5756–5764. [Google Scholar] [CrossRef]
- Wang, H.A.O.; Grolimund, D.; Giesen, C.; Borca, C.N.; Shaw-Stewart, J.R.H.; Bodenmiller, B.; Günther, D. Fast Chemical Imaging at High Spatial Resolution by Laser Ablation Inductively Coupled Plasma Mass Spectrometry. Anal. Chem. 2013, 85, 10107–10116. [Google Scholar] [CrossRef]
- Schoeberl, A.; Gutmann, M.; Theiner, S.; Schaier, M.; Schweikert, A.; Berger, W.; Koellensperger, G. Cisplatin Uptake in Macrophage Subtypes at the Single-Cell Level by LA-ICP-TOFMS Imaging. Anal. Chem. 2021, 93, 16456–16465. [Google Scholar] [CrossRef]
- Ali, H.R.; Jackson, H.W.; Zanotelli, V.R.T.; Danenberg, E.; Fischer, J.R.; Bardwell, H.; Provenzano, E.; CRUK IMAXT Grand Challenge Team; Rueda, O.M.; Chin, S.-F.; et al. Imaging Mass Cytometry and Multiplatform Genomics Define the Phenogenomic Landscape of Breast Cancer. Nat. Cancer 2020, 1, 163–175. [Google Scholar] [CrossRef]
- Giesen, C.; Wang, H.A.O.; Schapiro, D.; Zivanovic, N.; Jacobs, A.; Hattendorf, B.; Schüffler, P.J.; Grolimund, D.; Buhmann, J.M.; Brandt, S.; et al. Highly Multiplexed Imaging of Tumor Tissues with Subcellular Resolution by Mass Cytometry. Nat. Methods 2014, 11, 417–422. [Google Scholar] [CrossRef]
- Kuett, L.; Catena, R.; Özcan, A.; Plüss, A.; Ali, H.R.; Al Sa’d, M.; Alon, S.; Aparicio, S.; Battistoni, G.; Balasubramanian, S.; et al. Three-Dimensional Imaging Mass Cytometry for Highly Multiplexed Molecular and Cellular Mapping of Tissues and the Tumor Microenvironment. Nat. Cancer 2022, 3, 122–133. [Google Scholar] [CrossRef]
- Quatromoni, J.G.; Singhal, S.; Bhojnagarwala, P.; Hancock, W.W.; Albelda, S.M.; Eruslanov, E. An Optimized Disaggregation Method for Human Lung Tumors That Preserves the Phenotype and Function of the Immune Cells. J. Leukoc. Biol. 2015, 97, 201–209. [Google Scholar] [CrossRef] [Green Version]
- Han, G.; Spitzer, M.H.; Bendall, S.C.; Fantl, W.J.; Nolan, G.P. Metal-Isotope-Tagged Monoclonal Antibodies for High-Dimensional Mass Cytometry. Nat. Protoc. 2018, 13, 2121–2148. [Google Scholar] [CrossRef]
- Pace, H.E.; Rogers, N.J.; Jarolimek, C.; Coleman, V.A.; Higgins, C.P.; Ranville, J.F. Determining Transport Efficiency for the Purpose of Counting and Sizing Nanoparticles via Single Particle Inductively Coupled Plasma Mass Spectrometry. Anal. Chem. 2011, 83, 9361–9369. [Google Scholar] [CrossRef] [Green Version]
- Robinson, A.P.; Rodgers, J.M.; Goings, G.E.; Miller, S.D. Characterization of Oligodendroglial Populations in Mouse Demyelinating Disease Using Flow Cytometry: Clues for MS Pathogenesis. PLoS ONE 2014, 9, e107649. [Google Scholar] [CrossRef]
- Bajpai, R.; Lesperance, J.; Kim, M.; Terskikh, A.V. Efficient Propagation of Single Cells Accutase-Dissociated Human Embryonic Stem Cells. Mol. Reprod. Dev. 2008, 75, 818–827. [Google Scholar] [CrossRef]
- Corte-Rodríguez, M.; Blanco-González, E.; Bettmer, J.; Montes-Bayón, M. Quantitative Analysis of Transferrin Receptor 1 (TfR1) in Individual Breast Cancer Cells by Means of Labeled Antibodies and Elemental (ICP-MS) Detection. Anal. Chem. 2019, 91, 15532–15538. [Google Scholar] [CrossRef]
- Corte Rodríguez, M.; Álvarez-Fernández García, R.; Blanco, E.; Bettmer, J.; Montes-Bayón, M. Quantitative Evaluation of Cisplatin Uptake in Sensitive and Resistant Individual Cells by Single-Cell ICP-MS (SC-ICP-MS). Anal. Chem. 2017, 89, 11491–11497. [Google Scholar] [CrossRef]
- Marguerat, S.; Bähler, J. Coordinating Genome Expression with Cell Size. Trends Genet. 2012, 28, 560–565. [Google Scholar] [CrossRef]
- Bogdanos, D.P.; Gao, B.; Gershwin, M.E. Liver Immunology. Compr. Physiol. 2013, 3, 567–598. [Google Scholar] [PubMed] [Green Version]
- Thompson, C.B.; Scher, I.; Schaefer, M.E.; Lindsten, T.; Finkelman, F.D.; Mond, J.J. Size-Dependent B Lymphocyte Subpopulations: Relationship of Cell Volume to Surface Phenotype, Cell Cycle, Proliferative Response, and Requirements for Antibody Production to TNP-Ficoll and TNP-BA. J. Immunol. 1984, 133, 2333–2342. [Google Scholar] [PubMed]
- Cao, Y.; Feng, J.; Tang, L.; Yu, C.; Mo, G.; Deng, B. A Highly Efficient Introduction System for Single Cell- ICP-MS and Its Application to Detection of Copper in Single Human Red Blood Cells. Talanta 2020, 206, 120174. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, M.; Wang, B.; Zheng, L.; Chen, H.; Chai, Z.; Feng, W. Interrogating the Variation of Element Masses and Distribution Patterns in Single Cells Using ICP-MS with a High Efficiency Cell Introduction System. Anal. Bioanal. Chem. 2017, 409, 1415–1423. [Google Scholar] [CrossRef]
- Morrison, G.H.; Risby, T.H. Elemental Trace Analysis of Biological Materials. C R C Crit. Rev. Anal. Chem. 1979, 8, 287–320. [Google Scholar] [CrossRef]
- Cesta, M.F. Normal Structure, Function, and Histology of the Spleen. Toxicol. Pathol. 2006, 34, 455–465. [Google Scholar] [CrossRef]
- Ross, A.C.; Caballero, B.; Cousins, R.J.; Tucker, K.L.; Ziegler, T.R. Modern Nutrition in Health and Disease: Eleventh Edition; Wolters Kluwer Health Adis (ESP): Alphen aan den Rijn, The Netherlands, 2012. [Google Scholar]
- Hsu, M.Y.; Mina, E.; Roetto, A.; Porporato, P.E. Iron: An Essential Element of Cancer Metabolism. Cells 2020, 9, 2591. [Google Scholar] [CrossRef]
- Lou, X.; Zhang, G.; Herrera, I.; Kinach, R.; Ornatsky, O.; Baranov, V.; Nitz, M.; Winnik, M.A. Polymer-Based Elemental Tags for Sensitive Bioassays. Angew. Chem. Int. Ed. Engl. 2007, 46, 6111–6114. [Google Scholar] [CrossRef]
- Glass Expansion. Available online: https://www.geicp.com (accessed on 2 December 2022).





| Spleen | Liver | HepG2 | |
|---|---|---|---|
| Nd mass per cell (fg) | 0.088 ± 0.001 | 0.089 ± 0.001 | 0.231 ± 0.09 |
| TfR1 per cell | (1.86 ± 0.15) × 104 | (1.86 ± 0.17) × 104 | (4.97 ± 1.96) × 104 |
| Number of events | 279 | 195 | 202 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Álvarez-Fernández García, R.; Gutiérrez Romero, L.; Bettmer, J.; Montes-Bayón, M. Capabilities of Single Cell ICP-MS for the Analysis of Cell Suspensions from Solid Tissues. Nanomaterials 2023, 13, 12. https://doi.org/10.3390/nano13010012
Álvarez-Fernández García R, Gutiérrez Romero L, Bettmer J, Montes-Bayón M. Capabilities of Single Cell ICP-MS for the Analysis of Cell Suspensions from Solid Tissues. Nanomaterials. 2023; 13(1):12. https://doi.org/10.3390/nano13010012
Chicago/Turabian StyleÁlvarez-Fernández García, Roberto, Lucía Gutiérrez Romero, Jörg Bettmer, and Maria Montes-Bayón. 2023. "Capabilities of Single Cell ICP-MS for the Analysis of Cell Suspensions from Solid Tissues" Nanomaterials 13, no. 1: 12. https://doi.org/10.3390/nano13010012
APA StyleÁlvarez-Fernández García, R., Gutiérrez Romero, L., Bettmer, J., & Montes-Bayón, M. (2023). Capabilities of Single Cell ICP-MS for the Analysis of Cell Suspensions from Solid Tissues. Nanomaterials, 13(1), 12. https://doi.org/10.3390/nano13010012