Quantum Dot Doping-Induced Photoluminescence for Facile, Label-Free, and Sensitive Pyrophosphatase Activity Assay and Inhibitor Screening
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. Preparation of ZnSe QD
2.3. Fluorescent Assay for PPase
2.4. PPase inhibition Assay
2.5. Instruments
3. Results
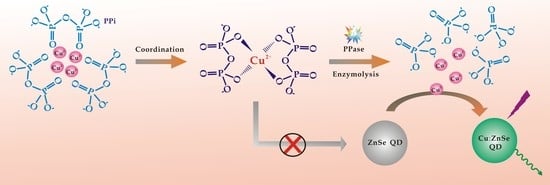
3.1. QD Photoluminescence Regulated by Cu2+/PPi
3.2. Optimization of the Experimental Conditions
3.3. Sensing Performance toward PPase Activity
3.4. PPase Inhibitor Screening
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Heikinheimo, P.; Lehtonen, J.; Baykov, A.; Lahti, R.; Cooperman, B.S.; Goldman, A. The structural basis for pyrophosphatase catalysis. Structure 1996, 4, 1491–1508. [Google Scholar] [CrossRef] [Green Version]
- Harold, F. Inorganic polyphosphates in biology: Structure, metabolism, and function. Bacteriol. Rev. 1966, 30, 772–794. [Google Scholar] [PubMed]
- Oksanen, E.; Ahonen, A.K.; Tuominen, H.; Tuominen, V.; Lahti, R.; Goldman, A.; Heikinheimo, P. A complete structural description of the catalytic cycle of yeast pyrophosphatase. Biochemistry 2007, 46, 1228–1239. [Google Scholar] [CrossRef] [PubMed]
- Carman, G.M.; Han, G.S. Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends Biochem. Sci. 2006, 31, 694–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lahti, R. Microbial inorganic pyrophosphatases. Microbiol. Rev. 1983, 47, 169–178. [Google Scholar] [PubMed]
- Ilias, M.; Young, T.W. Streptococcus gordonii soluble inorganic pyrophosphatase: An important role for the interdomain region in enzyme activity. Biochim. Biophys. Acta 2006, 1764, 1299–1306. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Hu, L.; Evers, S.; Chen, J.; Shen, Y. Differential expression profiling of human pancreatic adenocarcinoma and healthy pancreatic tissue. Proteomics 2004, 4, 3975–3988. [Google Scholar] [CrossRef]
- Koike, E.; Toda, S.; Yokoi, F.; Izuhara, K.; Koike, N.; Itoh, K.; Miyazaki, K.; Sugihara, H. Expression of new human inorganic pyrophosphatase in thyroid diseases: Its intimate association with hyperthyroidism. Biochem. Biophys. Res. Commun. 2006, 341, 691–696. [Google Scholar] [CrossRef]
- Friedman, D.; Hill, S.; Keller, J.; Merchant, N.; Levy, S.; Coffey, R.; Caprioli, R. Proteome analysis of human colon cancer by two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics 2004, 4, 793–811. [Google Scholar] [CrossRef]
- Cartier, P.H.; Thuillier, L. New radiochemical method for assay of inorganic pyrophosphatase. Anal. Biochem. 1971, 44, 397–403. [Google Scholar] [CrossRef]
- Eriksson, J.; Karamohamed, S.; Nyrén, P. Method for real-time detection of inorganic pyrophosphatase activity. Anal. Biochem. 2001, 293, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Vance, D.H.; Czarnik, A.W. Real-time assay of inorganic pyrophosphatase using a high-affinity chelation-enhanced fluorescence chemosensor. J. Am. Chem. Soc. 1994, 116, 9397–9398. [Google Scholar] [CrossRef]
- Deng, J.; Jiang, Q.; Wang, Y.; Yang, L.; Yu, P.; Mao, L. Real-time colorimetric assay of inorganic pyrophosphatase activity based on reversibly competitive coordination of Cu2+ between cysteine and pyrophosphate ion. Anal. Chem. 2013, 85, 9409–9415. [Google Scholar] [CrossRef]
- Sun, J.; Yang, F.; Zhao, D.; Yang, X. Highly sensitive real-time assay of inorganic pyrophosphatase activity based on the fluorescent gold nanoclusters. Anal. Chem. 2014, 86, 7883–7889. [Google Scholar] [CrossRef]
- Zhou, Q.; Lin, Y.; Xu, M.; Gao, Z.; Yang, H.; Tang, D. Facile synthesis of enhanced fluorescent gold–silver bimetallic nanocluster and its application for highly sensitive detection of inorganic pyrophosphatase activity. Anal. Chem. 2016, 88, 8886–8892. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Chen, Z.; Zhou, L.; Zheng, O.; Wu, X.; Guo, L.; Qiu, B.; Lin, Z.; Chen, G. Fluorometric method for inorganic pyrophosphatase activity detection and inhibitor screening based on click chemistry. Anal. Chem. 2015, 87, 816–820. [Google Scholar] [CrossRef]
- Sun, J.; Wang, B.; Zhao, X.; Li, Z.J.; Yang, X. Fluorescent and colorimetric dual-readout assay for inorganic pyrophosphatase with Cu2+-triggered oxidation of o-phenylenediamine. Anal. Chem. 2016, 88, 1355–1361. [Google Scholar] [CrossRef]
- Feng, X.; An, Y.; Yao, Z.; Li, C.; Shi, G. A turn-on fluorescent sensor for pyrophosphate based on the disassembly of Cu2+-mediated perylene diimide aggregates. ACS Appl. Mater. Interfaces 2012, 4, 614–618. [Google Scholar] [CrossRef]
- Ma, F.; Liu, W.; Tang, B.; Zhang, C. A single quantum dot-based nanosensor for the signal-on detection of DNA methyltransferase. Chem. Commun. 2017, 53, 6868–6871. [Google Scholar] [CrossRef]
- Silvi, S.; Credi, A. Luminescent sensors based on quantum dot–molecule conjugates. Chem. Soc. Rev. 2015, 44, 4275–4289. [Google Scholar] [CrossRef]
- Liang, G.X.; Li, L.L.; Liu, H.Y.; Zhang, J.R.; Burda, C.; Zhu, J.J. Fabrication of near-infrared-emitting CdSeTe/ZnS core/shell quantum dots and their electrogenerated chemiluminescence. Chem. Commun. 2010, 46, 2974–2976. [Google Scholar] [CrossRef] [PubMed]
- Gill, R.; Zayats, M.; Willner, I. Semiconductor quantum dots for bioanalysis. Angew. Chem. Int. Ed. 2008, 47, 7602–7625. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, N.; Goorskey, D.; Thessing, J.; Peng, X. An alternative of CdSe nanocrystal emitters: Pure and tunable impurity emissions in ZnSe nanocrystals. J. Am. Chem. Soc. 2005, 127, 17586–17587. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Baneyx, F. Aqueous, protein-driven synthesis of transition metal-doped ZnS immuno-quantum dots. ACS Nano 2011, 5, 8013–8018. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Shen, S.Z.; Sun, H.; Sun, K.; Liu, F. Synthesis of L-glutathione-capped-ZnSe quantum dots for the sensitive and selective determination of copper ion in aqueous solutions. Sens. Actuators B 2014, 203, 35–43. [Google Scholar] [CrossRef]
- Pang, J.; Lu, Y.; Gao, X.; Song, P.; Yang, F.; Liu, Y. On-off-on luminescent pyrophosphate probe based on the use of Mn-doped ZnS quantum dots and using Eu(III) as a mediator. Microchim. Acta 2018, 185, 480. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Ma, N. A general strategy for label-free sensitive DNA detection based on quantum dot doping. Anal. Chem. 2014, 86, 3676–3681. [Google Scholar] [CrossRef]
- Tian, Y.; Xin, C.; Liu, S.; Liu, Y.; Liu, S. Affinity binding-induced Hg2+ release and quantum dot doping for general, label-free, and homogenous fluorescence protein assay. ACS Sens. 2018, 3, 1401–1408. [Google Scholar] [CrossRef]
- Chen, C.; Zhao, D.; Sun, J.; Yang, X. Colorimetric logic gate for pyrophosphate and pyrophosphatase via regulating the catalytic capability of horseradish peroxidase. ACS Appl. Mater. Interfaces 2016, 8, 29529–29535. [Google Scholar] [CrossRef]
- Zhu, X.; Liu, J.; Peng, H.; Jiang, J.; Yu, R. A novel fluorescence assay for inorganic pyrophosphatase based on modulated aggregation of graphene quantum dots. Analyst 2016, 141, 251–255. [Google Scholar] [CrossRef]
- Ye, M.; Yu, Y.; Lin, B.; Cai, Y.; Cao, Y.; Guo, M.; Zhu, D. Copper nanoclusters reversible switches based on ions-triggered for detection of inorganic pyrophosphatase activity. Sens. Actuators B 2019, 284, 36–44. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, Y.; Liu, W.; Chu, L.; Liao, Z.; Guo, W.; Liu, G.Q.; He, X.; Wang, K. Electrochemical strategy for pyrophosphatase detection based on the peroxidase-like activity of G-quadruplex-Cu2+ DNAzyme. Talanta 2018, 178, 491–497. [Google Scholar] [CrossRef] [PubMed]





| Method | Detection Limit (mU/mL) | Linear Range (mU/mL) | Strategy | References |
|---|---|---|---|---|
| Colorimetry | 10 | 25.0–400.0 | Gold nanoparticles | [13] |
| Fluorescence | 0.03 | 0.1–30.0 | Au-Ag NCs | [15] |
| Fluorescence | 0.2 | 0.2–50.0 | o-Phenylenediamine oxidation | [17] |
| Fluorescence | 1.0 | 1.0–200.0 | Graphene quantum dots | [30] |
| Fluorescence | 1.3 | 3.0–40.0 | Copper nanoclusters | [31] |
| Eletrochemistry | 0.6 | 1.0–50.0 | G-quadruplex-Cu2+ DNAzyme | [32] |
| Fluorescence | 0.1 | 0.1–2.0 | Quantum dot doping | This work |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, Y.; Hao, L.; Wang, C.; Yang, X.; Liu, S. Quantum Dot Doping-Induced Photoluminescence for Facile, Label-Free, and Sensitive Pyrophosphatase Activity Assay and Inhibitor Screening. Nanomaterials 2019, 9, 111. https://doi.org/10.3390/nano9010111
Tian Y, Hao L, Wang C, Yang X, Liu S. Quantum Dot Doping-Induced Photoluminescence for Facile, Label-Free, and Sensitive Pyrophosphatase Activity Assay and Inhibitor Screening. Nanomaterials. 2019; 9(1):111. https://doi.org/10.3390/nano9010111
Chicago/Turabian StyleTian, Yishen, Lijie Hao, Chao Wang, Xiaoyan Yang, and Shufeng Liu. 2019. "Quantum Dot Doping-Induced Photoluminescence for Facile, Label-Free, and Sensitive Pyrophosphatase Activity Assay and Inhibitor Screening" Nanomaterials 9, no. 1: 111. https://doi.org/10.3390/nano9010111
APA StyleTian, Y., Hao, L., Wang, C., Yang, X., & Liu, S. (2019). Quantum Dot Doping-Induced Photoluminescence for Facile, Label-Free, and Sensitive Pyrophosphatase Activity Assay and Inhibitor Screening. Nanomaterials, 9(1), 111. https://doi.org/10.3390/nano9010111