Controlling the Self-Assembly of Biomolecules into Functional Nanomaterials through Internal Interactions and External Stimulations: A Review
Abstract
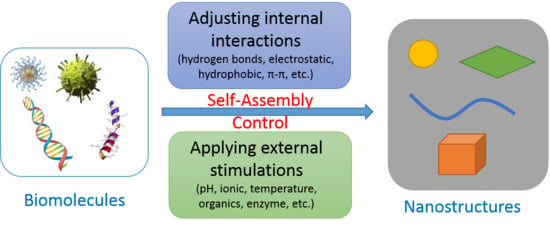
:1. Introduction
2. Internal Interactions towards Biomolecular Self-Assembly
2.1. Basic Molecular Interactions
2.1.1. Hydrogen Bonds
2.1.2. Electrostatic Interaction
2.1.3. Hydrophobic Interaction
2.1.4. π–π Interaction
2.2. Biomolecular-Specific Interactions
2.2.1. DNA/RNA Base Pairing
2.2.2. Ligand–Receptor Binding
2.2.3. Biomolecule–Polymer Conjugates for Self-Assembly
3. External Stimulations towards Biomolecular Self-Assembly
3.1. pH Effect
3.2. Temperature Effect
3.3. Ionic Effect
3.4. Organic Stimulators
3.5. Enzymatic Stimulators
3.6. Photo-Stimulation
3.7. Tailoring Molecular Structure
4. Various Self-Assembled Biological Nanostructures/Materials
5. Conclusions and Outlooks
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Gwo, S.; Chen, H.Y.; Lin, M.H.; Sun, L.Y.; Li, X.Q. Nanomanipulation and controlled self-assembly of metal nanoparticles and nanocrystals for plasmonics. Chem. Soc. Rev. 2016, 45, 5672–5716. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, Y.J.; Li, Z.; Wu, A.G.; Wei, G. Bottom-up synthesis and sensor applications of biomimetic nanostructures. Materials 2016, 9, 53. [Google Scholar] [CrossRef]
- Hoheisel, T.N.; Hur, K.; Wiesner, U.B. Block copolymer-nanoparticle hybrid self-assembly preface. Prog. Polym. Sci. 2015, 40, 3–32. [Google Scholar] [CrossRef]
- Bhattacharyya, K.; Mukherjee, S. Fluorescent metal nano-clusters as next generation fluorescent probes for cell imaging and drug delivery. Bull. Chem. Soc. Jpn. 2018, 91, 447–454. [Google Scholar] [CrossRef]
- Komiyama, M.; Mori, T.; Ariga, K. Molecular imprinting: Materials nanoarchitectonics with molecular information. Bull. Chem. Soc. Jpn. 2018, 91, 1075–1111. [Google Scholar] [CrossRef]
- Rogers, W.B.; Shih, W.M.; Manoharan, V.N. Using DNA to program the self-assembly of colloidal nanoparticles and microparticles. Nat. Rev. Mater. 2016, 1, 16008. [Google Scholar] [CrossRef]
- Bai, Y.S.; Luo, Q.; Liu, J.Q. Protein self-assembly via supramolecular strategies. Chem. Soc. Rev. 2016, 45, 2756–2767. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Su, Z.Q.; Reynolds, N.P.; Arosio, P.; Hamley, I.W.; Gazit, E.; Mezzenga, R. Self-assembling peptide and protein amyloids: From structure to tailored function in nanotechnology. Chem. Soc. Rev. 2017, 46, 4661–4708. [Google Scholar] [CrossRef]
- Zhang, W.S.; Yu, X.Q.; Li, Y.; Su, Z.Q.; Jandt, K.D.; Wei, G. Protein-mimetic peptide nanofibers: Motif design, self-assembly synthesis, and sequence-specific biomedical applications. Prog. Polym. Sci. 2018, 80, 94–124. [Google Scholar] [CrossRef]
- Milles, S.; Jensen, M.R.; Communie, G.; Maurin, D.; Schoehn, G.; Ruigrok, R.W.H.; Blackledge, M. Self-assembly of measles virus nucleocapsid-like particles: Kinetics and rna sequence dependence. Angew. Chem. Int. Ed. 2016, 55, 9356–9360. [Google Scholar] [CrossRef]
- Sawada, T.; Serizawa, T. Filamentous viruses as building blocks for hierarchical self-assembly toward functional soft materials. Bull. Chem. Soc. Jpn. 2018, 91, 455–466. [Google Scholar] [CrossRef]
- Zhou, J.; Du, X.W.; Berciu, C.; He, H.J.; Shi, J.F.; Nicastro, D.; Xu, B. Enzyme-instructed self-assembly for spatiotemporal profiling of the activities of alkaline phosphatases on live cells. Chem 2016, 1, 246–263. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.M.; Feng, Z.Q.Q.; Wang, Y.Z.; Zhou, R.; Yang, Z.M.; Xu, B. Integrating enzymatic self-assembly and mitochondria targeting for selectively killing cancer cells without acquired drug resistance. J. Am. Chem. Soc. 2016, 138, 16046–16055. [Google Scholar] [CrossRef]
- Lin, Y.Y.; Chapman, R.; Stevens, M.M. Integrative self-assembly of graphene quantum dots and biopolymers into a versatile biosensing toolkit. Adv. Funct. Mater. 2015, 25, 3183–3192. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Cao, Y.Y.; Huang, Z.H.; Duan, Y.Y.; Che, S.N. Silica biomineralization via the self-assembly of helical biomolecules. Adv. Mater. 2015, 27, 479–497. [Google Scholar] [CrossRef] [PubMed]
- Stephanopoulos, N.; Ortony, J.H.; Stupp, S.I. Self-assembly for the synthesis of functional biomaterials. Acta Mater. 2013, 61, 912–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mauro, M.; Aliprandi, A.; Septiadi, D.; Kehra, N.S.; De Cola, L. When self-assembly meets biology: Luminescent platinum complexes for imaging applications. Chem. Soc. Rev. 2014, 43, 4144–4166. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.Z.; Hu, R.; Zhao, Z.L.; Chen, Z.; Zhang, X.B.; Tan, W.H. Noncanonical self-assembly of multifunctional DNA nanoflowers for biomedical applications. J. Am. Chem. Soc. 2013, 135, 16438–16445. [Google Scholar] [CrossRef]
- Wang, L.; Wu, A.G.; Wei, G. Graphene-based aptasensors: From molecule-interface interactions to sensor design and biomedical diagnostics. Analyst 2018, 143, 1526–1543. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Y.J.; Wu, A.G.; Wei, G. Designed graphene-peptide nanocomposites for biosensor applications: A review. Anal. Chim. Acta 2017, 985, 24–40. [Google Scholar] [CrossRef]
- Li, Q.; Jia, Y.; Dai, L.R.; Yang, Y.; Li, J.B. Controlled rod nanostructured assembly of diphenylalanine and their optical waveguide properties. ACS Nano 2015, 9, 2689–2695. [Google Scholar] [CrossRef] [PubMed]
- Caplan, M.R.; Moore, P.N.; Zhang, S.G.; Kamm, R.D.; Lauffenburger, D.A. Self-assembly of a beta-sheet protein governed by relief of electrostatic repulsion relative to van der waals attraction. Biomacromolecules 2000, 1, 627–631. [Google Scholar] [CrossRef] [PubMed]
- Elsawy, M.A.; Smith, A.M.; Hodson, N.; Squires, A.; Miller, A.F.; Saiani, A. Modification of beta-sheet forming peptide hydrophobic face: Effect on self-assembly and gelation. Langmuir 2016, 32, 4917–4923. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.B.; Chao, J.; Liu, H.J.; Su, S.; Wang, L.H.; Huang, W.; Willner, I.; Fan, C.H. Clamped hybridization chain reactions for the self-assembly of patterned DNA hydrogels. Angew. Chem. Int. Ed. 2017, 56, 2171–2175. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Reichert, J.; Bossert, J.; Jandt, K.D. Novel biopolymeric template for the nucleation and growth of hydroxyapatite crystals based on self-assembled fibrinogen fibrils. Biomacromolecules 2008, 9, 3258–3267. [Google Scholar] [CrossRef]
- Wei, G.; Reichert, J.; Jandt, K.D. Controlled self-assembly and templated metallization of fibrinogen nanofibrils. Chem. Commun. 2008, 3903–3905. [Google Scholar] [CrossRef]
- Dave, A.C.; Loveday, S.M.; Anema, S.G.; Jameson, G.B.; Singh, H. Modulating beta-lactoglobulin nanofibril self-assembly at pH 2 using glycerol and sorbitol. Biomacromolecules 2014, 15, 95–103. [Google Scholar] [CrossRef]
- Li, R.; Horgan, C.C.; Long, B.; Rodriguez, A.L.; Mather, L.; Barrow, C.J.; Nisbet, D.R.; Williams, R.J. Tuning the mechanical and morphological properties of self-assembled peptide hydrogels via control over the gelation mechanism through regulation of ionic strength and the rate of ph change. RSC Adv. 2015, 5, 301–307. [Google Scholar] [CrossRef]
- Kim, W.; Thevenot, J.; Ibarboure, E.; Lecommandoux, S.; Chaikof, E.L. Self-assembly of thermally responsive amphiphilic diblock copolypeptides into spherical micellar nanoparticles. Angew. Chem. Int. Ed. 2010, 49, 4257–4260. [Google Scholar] [CrossRef]
- Yu, X.L.; Du, R.F.; Li, B.Y.; Zhang, Y.H.; Liu, H.J.; Qu, J.H.; An, X.Q. Biomolecule-assisted self-assembly of cds/mos2/graphene hollow spheres as high-efficiency photocatalysts for hydrogen evolution without noble metals. Appl. Catal. B 2016, 182, 504–512. [Google Scholar] [CrossRef]
- Zhao, Z.; Liu, Y.; Yan, H. DNA origami templated self-assembly of discrete length single wall carbon nanotubes. Org. Biomol. Chem. 2013, 11, 596–598. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Zhang, Y.; Steckbeck, S.; Su, Z.Q.; Li, Z. Biomimetic graphene-fept nanohybrids with high solubility, ferromagnetism, fluorescence, and enhanced electrocatalytic activity. J. Mater. Chem. 2012, 22, 17190–17195. [Google Scholar] [CrossRef]
- Wang, J.H.; Ouyang, Z.F.; Ren, Z.W.; Li, J.F.; Zhang, P.P.; Wei, G.; Su, Z.Q. Self-assembled peptide nanofibers on graphene oxide as a novel nanohybrid for biomimetic mineralization of hydroxyapatite. Carbon 2015, 89, 20–30. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, M.X.; Zhu, C.Y.; Nurumbetov, G.; Li, Z.D.; Wilson, P.; Kempe, K.; Haddleton, D.M. Well-defined protein/peptide-polymer conjugates by aqueous cu-lrp: Synthesis and controlled self-assembly. J. Am. Chem. Soc. 2015, 137, 9344–9353. [Google Scholar] [CrossRef] [PubMed]
- Zou, Q.L.; Abbas, M.; Zhao, L.Y.; Li, S.K.; Shen, G.Z.; Yan, X.H. Biological photothermal nanodots based on self-assembly of peptide porphyrin conjugates for antitumor therapy. J. Am. Chem. Soc. 2017, 139, 1921–1927. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.G. Fabrication of novel biomaterials through molecular self-assembly. Nat. Biotechnol. 2003, 21, 1171–1178. [Google Scholar] [CrossRef] [PubMed]
- Groeger, C.; Lutz, K.; Brunner, E. Biomolecular self-assembly and its relevance in silica biomineralization. Cell Biochem. Biophys. 2008, 50, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.G.; Marini, D.M.; Hwang, W.; Santoso, S. Design of nanostructured biological materials through self-assembly of peptides and proteins. Curr. Opin. Chem. Biol. 2002, 6, 865–871. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.L.; Liu, A.J.; Cao, S.Q.; Putri, R.M.; Jonkheijm, P.; Cornelissen, J.J.L.M. Self-assembly of proteins: Towards supramolecular materials. Chem. Eur. J. 2016, 22, 15570–15582. [Google Scholar] [CrossRef]
- Willner, I.; Willner, B. Biomolecule-based nanomaterials and nanostructures. Nano Lett. 2010, 10, 3805–3815. [Google Scholar] [CrossRef]
- McManus, J.J.; Charbonneau, P.; Zaccarelli, E.; Asherie, N. The physics of protein self-assembly. Curr. Opin. Colloid Interface Sci. 2016, 22, 73–79. [Google Scholar] [CrossRef] [Green Version]
- Qi, G.B.; Gao, Y.J.; Wang, L.; Wang, H. Self-assembled peptide-based nanomaterials for biomedical imaging and therapy. Adv. Mater. 2018, 30, 1703444. [Google Scholar] [CrossRef]
- Marchesan, S.; Vargiu, A.V.; Styan, K.E. The phe-phe motif for peptide self-assembly in nanomedicine. Molecules 2015, 20, 19775–19788. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, D.; Kiselev, M.A.; Magazu, S.; Calandra, P. Amphiphiles self-assembly: Basic concepts and future perspectives of supramolecular approaches. Adv. Cond. Matter. Phys. 2015, 2015, 151683. [Google Scholar] [CrossRef]
- Wang, Z.G.; Ding, B.Q. Engineering DNA self-assemblies as templates for functional nanostructures. Acc. Chem. Res. 2014, 47, 1654–1662. [Google Scholar] [CrossRef] [PubMed]
- Shaham-Niv, S.; Adler-Abramovich, L.; Schnaider, L.; Gazit, E. Extension of the generic amyloid hypothesis to nonproteinaceous metabolite assemblies. Sci. Adv. 2015, 1, e1500137. [Google Scholar] [CrossRef]
- Shaham-Niv, S.; Arnon, Z.A.; Sade, D.; Lichtenstein, A.; Shirshin, E.A.; Kolusheva, S.; Gazit, E. Intrinsic fluorescence of metabolite amyloids allows label-free monitoring of their formation and dynamics in live cell. Angew. Chem. Int. Ed. 2018, 57, 12444–12447. [Google Scholar] [CrossRef]
- Shaham-Niv, S.; Rehak, P.; Zaguri, D.; Kolusheva, S.; Kral, P.; Gazit, E. Metabolite amyloid-like fibrils interact with model membranes. Chem. Commun. 2018, 54, 4561–4564. [Google Scholar] [CrossRef] [PubMed]
- Bartocci, S.; Berrocal, J.A.; Guarracino, P.; Grillaud, M.; Franco, L.; Mba, M. Peptide-driven charge-transfer organogels built from synergetic hydrogen bonding and pyrene-naphthalenediimide donor-acceptor interactions. Chem. Eur. J. 2018, 24, 2920–2928. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.L.; Bobbitt, N.S.; Logsdon, J.L.; Powers-Riggs, N.E.; Nelson, J.N.; Liu, X.L.; Wang, T.C.; Snurr, R.Q.; Hupp, J.T.; Farha, O.K.; et al. Tunable crystallinity and charge transfer in two-dimensional g-quadruplex organic frameworks. Angew. Chem. Int. Ed. 2018, 57, 3985–3989. [Google Scholar] [CrossRef]
- Bilbao, N.; Destoop, I.; De Feyter, S.; Gonzalez-Rodriguez, D. Two-dimensional nanoporous networks formed by liquid-to-solid transfer of hydrogen-bonded macrocycles built from DNA bases. Angew. Chem. Int. Ed. 2016, 55, 659–663. [Google Scholar] [CrossRef]
- Li, P.P.; Chen, X.; Yang, W.S. Graphene-induced self-assembly of peptides into macroscopic-scale organized nanowire arrays for electrochemical nadh sensing. Langmuir 2013, 29, 8629–8635. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.M.; Jung, B.; Kim, Y.H.; Park, A.R.; Han, S.; Choe, W.S.; Yoo, P.J. Nanomesh-structured ultrathin membranes harnessing the unidirectional alignment of viruses on a graphene-oxide film. Adv. Mater. 2014, 26, 3899–3904. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.Y.; Cui, X.Q.; Guan, W.M.; Wang, Q.Y.; Liu, C.; Wang, H.T.; Qi, K.; Singh, D.J.; Zheng, W.T. Surface plasmon resonance technique for directly probing the interaction of DNA and graphene oxide and ultra-sensitive biosensing. Biosens. Bioelectron. 2014, 58, 374–379. [Google Scholar] [CrossRef] [PubMed]
- Kundu, B.; Eltohamy, M.; Yadavalli, V.K.; Kundu, S.C.; Kim, H.W. Biomimetic designing of functional silk nanotopography using self assembly. ACS Appl. Mater. Interfaces 2016, 8, 28458–28467. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Fan, Q.S.; Zhao, L.L.; Qiao, Q.L.; Zhang, X.Y.; Hou, C.X.; Xu, J.Y.; Luo, Q.; Liu, J.Q. The construction of functional protein nanotubes by small molecule-induced self-assembly of cricoid proteins. Chem. Commun. 2016, 52, 4092–4095. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.D.; Chen, L.; Ding, X.F.; Xu, L.D.; Zhou, X.R.; Wei, P.; Liang, J.F.; Luo, S.Z. High-resolution insights into the stepwise self-assembly of nanofiber from bioactive peptides. J. Phys. Chem. B 2017, 121, 7421–7430. [Google Scholar] [CrossRef]
- Wang, J.H.; Zhao, X.J.; Li, J.F.; Kuang, X.; Fan, Y.Q.; Wei, G.; Su, Z.Q. Electrostatic assembly of peptide nanofiber-biomimetic silver nanowires onto graphene for electrochemical sensors. ACS Macro Lett. 2014, 3, 529–533. [Google Scholar] [CrossRef]
- Palchoudhury, S.; Zhou, Z.Y.; Ramasamy, K.; Okirie, F.; Prevelige, P.E.; Gupta, A. Self-assembly of p22 protein cages with polyamidoamine dendrimer and inorganic nanoparticles. J. Mater. Res. 2017, 32, 465–472. [Google Scholar] [CrossRef]
- Sun, H.C.; Zhang, X.Y.; Miao, L.; Zhao, L.L.; Luo, Q.; Xu, J.Y.; Liu, J.Q. Micelle-induced self-assembling protein nanowires: Versatile supramolecular scaffolds for designing the light-harvesting system. ACS Nano 2016, 10, 421–428. [Google Scholar] [CrossRef]
- Zhao, L.L.; Zou, H.Y.; Zhang, H.; Sun, H.C.; Wang, T.T.; Pan, T.Z.; Li, X.M.; Bai, Y.S.; Ojao, S.P.; Luo, Q.; et al. Enzyme-triggered defined protein nanoarrays: Efficient light-harvesting systems to mimic chloroplasts. ACS Nano 2017, 11, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.H.; Li, Z.Y.; Rong, L.; Qin, S.Y.; Lei, Q.; Cheng, H.; Zhou, X.; Zhuo, R.X.; Zhang, X.Z. Self-assembly of hybridized peptide nucleic acid amphiphiles. ACS Macro Lett. 2014, 3, 467–471. [Google Scholar] [CrossRef]
- McGuinness, K.; Nanda, V. Collagen mimetic peptide discs promote assembly of a broad range of natural protein fibers through hydrophobic interactions. Org. Biomol. Chem. 2017, 15, 5893–5898. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Abiko, S.; Himeiwa, T.; Kinoshita, T. Two-dimensional self-assembly of amphiphilic peptides; adsorption-induced secondary structural transition on hydrophilic substrate. J. Colloid Interface Sci. 2015, 442, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.S.; Lin, J.; Liu, Y.; Huang, P.; Jin, A.; Chen, X.Y. Self-assembly mechanisms of nanofibers from peptide amphiphiles in solution and on substrate surfaces. Nanoscale 2016, 8, 14814–14820. [Google Scholar] [CrossRef]
- Yang, H.; Fung, S.Y.; Pritzker, M.; Chen, P. Modification of hydrophilic and hydrophobic surfaces using an ionic-complementary peptide. PLOS One 2007, 2, e1325. [Google Scholar] [CrossRef] [PubMed]
- Anand, B.G.; Dubey, K.; Shekhawat, D.S.; Prajapati, K.P.; Kar, K. Strategically designed antifibrotic gold nanoparticles to prevent collagen fibril formation. Langmuir 2017, 33, 13252–13261. [Google Scholar] [CrossRef]
- Wang, J.Q.; Tao, K.; Yang, Y.Z.; Zhang, L.Y.; Wang, D.; Cao, M.W.; Sun, Y.W.; Xia, D.H. Short peptide mediated self-assembly of platinum nanocrystals with selective spreading property. RSC Adv. 2016, 6, 58099–58105. [Google Scholar] [CrossRef]
- Li, Q.; Liu, L.; Zhang, S.; Xu, M.; Wang, X.Q.; Wang, C.; Besenbacher, F.; Dong, M.D. Modulating a beta(33-42) peptide assembly by graphene oxide. Chem. Eur. J. 2014, 20, 7236–7240. [Google Scholar] [CrossRef]
- Wang, E.; Desai, M.S.; Lee, S.W. Light-controlled graphene-elastin composite hydrogel actuators. Nano Lett. 2013, 13, 2826–2830. [Google Scholar] [CrossRef]
- 71 Lu, C.H.; Li, J.; Zhang, X.L.; Zheng, A.X.; Yang, H.H.; Chen, X.; Chen, G.N. General approach for monitoring peptide-protein interactions based on graphene-peptide complex. Anal. Chem. 2011, 83, 7276–7282. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Liu, X.M.; Wu, G.Y.; Zhou, P.; Zhou, Y.T.; Wang, L.; Huang, X. Efficient way to generate protein-based nanoparticles by in-situ photoinitiated polymerization-induced self-assembly. ACS Macro Lett. 2017, 6, 689–694. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.Y.; Huang, X.L.; Zhou, X.J.; Wu, H.X.; Guo, S.W. Assembly of graphene oxide-enzyme conjugates through hydrophobic interaction. Small 2012, 8, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Law, A.S.Y.; Yeung, M.C.L.; Yam, V.W.W. Arginine-rich peptide-induced supramolecular self-assembly of water-soluble anionic alkynylplatinum(ii) complexes: A continuous and label-free luminescence assay for trypsin and inhibitor screening. ACS Appl. Mater. Interfaces 2017, 9, 41143–41150. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.K.; Lin, Z.H.; Liu, Q.; Jiang, S.; Liu, H.; Su, X.G. DNA-hosted copper nanoclusters/graphene oxide based fluorescent biosensor for protein kinase activity detection. Anal. Chim. Acta 2018, 1012, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.Q.; Shen, H.Y.; Wang, H.X.; Wang, J.H.; Li, J.F.; Nienhaus, G.U.; Shang, L.; Wei, G. Motif-designed peptide nanofibers decorated with graphene quantum dots for simultaneous targeting and imaging of tumor cells. Adv. Funct. Mater. 2015, 25, 5472–5478. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, W.S.; Zhang, L.; Li, J.F.; Su, Z.Q.; Wei, G. Sequence-designed peptide nanofibers bridged conjugation of graphene quantum dots with graphene oxide for high performance electrochemical hydrogen peroxide biosensor. Adv. Mater. Interfaces 2017, 4, 1600895. [Google Scholar] [CrossRef]
- Li, D.P.; Zhang, W.S.; Yu, X.Q.; Wang, Z.P.; Su, Z.Q.; Wei, G. When biomolecules meet graphene: From molecular level interactions to material design and applications. Nanoscale 2016, 8, 19491–19509. [Google Scholar] [CrossRef]
- Yun, W.; Xiong, W.; Wu, H.; Fu, M.; Huang, Y.; Liu, X.Y.; Yang, L.Z. Graphene oxide-based fluorescent “turn-on” strategy for hg2+ detection by using catalytic hairpin assembly for amplification. Sens. Actuat. B 2017, 249, 493–498. [Google Scholar] [CrossRef]
- Wang, J.M.; Zhu, H.H.; Xu, Y.H.; Yang, W.R.; Liu, A.; Shan, F.K.; Cao, M.M.; Liu, J.Q. Graphene nanodots encaged 3-d gold substrate as enzyme loading platform for the fabrication of high performance biosensors. Sens. Actuat. B 2015, 220, 1186–1195. [Google Scholar] [CrossRef]
- Huang, J.Y.; Zhao, L.; Lei, W.; Wen, W.; Wang, Y.J.; Bao, T.; Xiong, H.Y.; Zhang, X.H.; Wang, S.F. A high-sensitivity electrochemical aptasensor of carcinoembryonic antigen based on graphene quantum dots-ionic liquid-nafion nanomatrix and dnazyme-assisted signal amplification strategy. Biosens. Bioelectron. 2018, 99, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Li, M.Z.; Pan, Y.X.; Guo, X.Y.; Liang, Y.H.; Wu, Y.P.; Wen, Y.; Yang, H.F. Pt/single-stranded DNA/graphene nanocomposite with improved catalytic activity and co tolerance. J. Mater. Chem. A 2015, 3, 10353–10359. [Google Scholar] [CrossRef]
- Shen, W.L.; Liu, Q.; Ding, B.Q.; Zhu, C.Q.; Shen, Z.Y.; Seeman, N.C. Facilitation of DNA self-assembly by relieving the torsional strains between building blocks. Org. Biomol. Chem. 2017, 15, 465–469. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Yang, J.; Jiang, S.X.; Liu, Y.; Yan, H. Dnazyme-based logic gate-mediated DNA self-assembly. Nano Lett. 2016, 16, 736–741. [Google Scholar] [CrossRef]
- Elbaz, J.; Yin, P.; Voigt, C.A. Genetic encoding of DNA nanostructures and their self-assembly in living bacteria. Nat. Commun. 2016, 7, 11179. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Yan, H. DNA self-assembly scaled up. Nature 2017, 552, 185. [Google Scholar] [CrossRef]
- Li, Z.; Liu, M.H.; Wang, L.; Nangreave, J.; Yan, H.; Liu, Y. Molecular behavior of DNA origami in higher-order self-assembly. J. Am. Chem. Soc. 2010, 132, 13545–13552. [Google Scholar] [CrossRef] [PubMed]
- Idili, A.; Vallee-Belisle, A.; Ricci, F. Programmable pH-triggered DNA nanoswitches. J. Am. Chem. Soc. 2014, 136, 5836–5839. [Google Scholar] [CrossRef]
- Kuzyk, A.; Laitinen, K.T.; Torma, P. DNA origami as a nanoscale template for protein assembly. Nanotechnology 2009, 20, 235305. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Deng, Z.T.; Ding, B.Q.; Yan, H.; Liu, Y. DNA-origami-directed self-assembly of discrete silver-nanoparticle architectures. Angew. Chem. Int. Ed. 2010, 49, 2700–2704. [Google Scholar] [CrossRef]
- Li, H.; Zhang, K.M.; Pi, F.M.; Guo, S.J.; Shlyakhtenko, L.; Chiu, W.; Shu, D.; Guo, P.X. Controllable self-assembly of rna tetrahedrons with precise shape and size for cancer targeting. Adv. Mater. 2016, 28, 7501–7507. [Google Scholar] [CrossRef] [PubMed]
- Boerneke, M.A.; Dibrov, S.M.; Hermann, T. Crystal-structure-guided design of self-assembling RNA nanotriangles. Angew. Chem. Int. Ed. 2016, 55, 4097–4100. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.M.; Subramanian, H.K.K.; Franco, E. Self-assembly of multi-stranded rna motifs into lattices and tubular structures (vol 19, pg 5449, 2017). Nucleic Acids Res. 2017, 45, 5628. [Google Scholar] [CrossRef] [PubMed]
- Berger, O.; Adler-Abramovich, L.; Levy-Sakin, M.; Grunwald, A.; Liebes-Peer, Y.; Bachar, M.; Buzhansky, L.; Mossou, E.; Forsyth, V.T.; Schwartz, T.; et al. Light-emitting self-assembled peptide nucleic acids exhibit both stacking interactions and watson-crick base pairing. Nat. Nanotechnol. 2015, 10, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Liljestrom, V.; Mikkila, J.; Kostiainen, M.A. Self-assembly and modular functionalization of three-dimensional crystals from oppositely charged proteins. Nat. Commun. 2014, 5, 4445. [Google Scholar] [CrossRef] [PubMed]
- Haburcak, R.; Shi, J.F.; Du, X.W.; Yuan, D.; Xu, B. Ligand-receptor interaction modulates the energy landscape of enzyme-instructed self-assembly of small molecules. J. Am. Chem. Soc. 2016, 138, 15397–15404. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, S.J.; Petitzon, M.; Mognetti, B.M. Bond formation kinetics affects self-assembly directed by ligand-receptor interactions. Soft Matter 2016, 12, 9585–9592. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.P.; Mamedova, N.; Kotov, N.A.; Chen, W.; Studer, J. Antigen/antibody immunocomplex from cdte nanoparticle bioconjugates. Nano Lett. 2002, 2, 817–822. [Google Scholar] [CrossRef]
- Kominami, H.; Kobayashi, K.; Ido, S.; Kimiya, H.; Yamada, H. Immunoactivity of self-assembled antibodies investigated by atomic force microscopy. RSC Adv. 2018, 8, 29378–29384. [Google Scholar] [CrossRef]
- Fong, W.K.; Negrini, R.; Vallooran, J.J.; Mezzenga, R.; Boyd, B.J. Responsive self-assembled nanostructured lipid systems for drug delivery and diagnostics. J. Colloid Interface Sci. 2016, 484, 320–339. [Google Scholar] [CrossRef]
- Zhou, Y.X.; Briand, V.A.; Sharma, N.; Ahn, S.K.; Kasi, R.M. Polymers comprising cholesterol: Synthesis, self-assembly, and applications. Materials 2009, 2, 636–660. [Google Scholar] [CrossRef]
- Conn, C.E.; Drummond, C.J. Nanostructured bicontinuous cubic lipid self-assembly materials as matrices for protein encapsulation. Soft Matter 2013, 9, 3449–3464. [Google Scholar] [CrossRef]
- Zerkoune, L.; Lesieur, S.; Putaux, J.L.; Choisnard, L.; Geze, A.; Wouessidjewe, D.; Angelov, B.; Vebert-Nardin, C.; Doutch, J.; Angelova, A. Mesoporous self-assembled nanoparticles of biotransesterified cyclodextrins and nonlamellar lipids as carriers of water-insoluble substances. Soft Matter 2016, 12, 7539–7550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.K.; Ren, L.J.; Wu, H.; Wang, W. Self-assembly of the polyoxometalate-cholesterol conjugate into microrods or nanoribbons regulated by thermodynamics. New J. Chem. 2016, 40, 954–961. [Google Scholar] [CrossRef]
- Engberg, K.; Waters, D.J.; Kelmanovich, S.; Parke-Houben, R.; Hartmann, L.; Toney, M.F.; Frank, C.W. Self-assembly of cholesterol tethered within hydrogel networks. Polymer 2016, 84, 371–382. [Google Scholar] [CrossRef] [Green Version]
- Lei, H.R.; Liu, J.; Yan, J.L.; Quan, J.M.; Fang, Y. Luminescent helical nanofiber self-assembled from a cholesterol-based metalloamphiphile and its application in DNA conformation recognition. Langmuir 2016, 32, 10350–10357. [Google Scholar] [CrossRef] [PubMed]
- Silva, N.H.C.S.; Pinto, R.J.B.; Freire, C.S.R.; Marrucho, I.M. Production of lysozyme nanofibers using deep eutectic solvent aqueous solutions. Colloids Surf. B 2016, 147, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Dinesh, B.; Squillaci, M.A.; Menard-Moyon, C.; Samori, P.; Bianco, A. Self-assembly of diphenylalanine backbone homologues and their combination with functionalized carbon nanotubes. Nanoscale 2015, 7, 15873–15879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moyer, T.J.; Finbloom, J.A.; Chen, F.; Toft, D.J.; Cryns, V.L.; Stupp, S.I. pH and amphiphilic structure direct supramolecular behavior in biofunctional assemblies. J. Am. Chem. Soc. 2014, 136, 14746–14752. [Google Scholar] [CrossRef] [PubMed]
- Cote, Y.; Fu, I.W.; Dobson, E.T.; Goldberger, J.E.; Nguyen, H.D.; Shen, J.K. Mechanism of the pH-controlled self-assembly of nanofibers from peptide amphiphiles. J. Phys. Chem. C 2014, 118, 16272–16278. [Google Scholar] [CrossRef]
- Jana, P.; Ehlers, M.; Zellermann, E.; Samanta, K.; Schmuck, C. Ph-controlled formation of a stable beta-sheet and amyloid-like fibers from an amphiphilic peptide: The importance of a tailor-made binding motif for secondary structure formation. Angew. Chem. Int. Ed. 2016, 55, 15287–15291. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, M.C.; Liang, C.; Mehta, A.K.; Lynn, D.G.; Grover, M.A. Multistep conformation selection in amyloid assembly. J. Am. Chem. Soc. 2017, 139, 17007–17010. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Haverick, M.; Stump, K.; Yang, X.Y.; Tweedle, M.F.; Goldberger, J.E. Fine-tuning the pH trigger of self-assembly. J. Am. Chem. Soc. 2012, 134, 3647–3650. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.R.; Gan, H.X.; Tong, Y.W. pH-controlled hierarchical self-assembly of peptide amphiphile. Macromolecules 2015, 48, 2647–2653. [Google Scholar] [CrossRef]
- Larnaudie, S.C.; Brendel, J.C.; Jolliffe, K.A.; Perrier, S. pH-responsive, amphiphilic core-shell supramolecular polymer brushes from cyclic peptide-polymer conjugates. ACS Macro Lett. 2017, 6, 1347–1351. [Google Scholar] [CrossRef]
- Reinecke, A.; Brezesinski, G.; Harrington, M.J. pH-responsive self-organization of metal-binding protein motifs from biomolecular junctions in mussel byssus. Adv. Mater. Interfaces 2017, 4, 1600416. [Google Scholar] [CrossRef]
- Brodin, J.D.; Ambroggio, X.I.; Tang, C.Y.; Parent, K.N.; Baker, T.S.; Tezcan, F.A. Metal-directed, chemically tunable assembly of one-, two- and three-dimensional crystalline protein arrays. Nat. Chem. 2012, 4, 375–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brodin, J.D.; Carr, J.R.; Sontz, P.A.; Tezcan, F.A. Exceptionally stable, redox-active supramolecular protein assemblies with emergent properties. Proc. Natl. Acad. Sci. USA 2014, 111, 2897–2902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elbaz, J.; Wang, Z.G.; Orbach, R.; Willner, I. pH-stimulated concurrent mechanical activation of two DNA “tweezers”. A “set-reset” logic gate system. Nano Lett. 2009, 9, 4510–4514. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.H.; Cecconello, A.; Elbaz, J.; Credi, A.; Willner, I. A three-station DNA catenane rotary motor with controlled directionality. Nano Lett. 2013, 13, 2303–2308. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Willner, I. pH-stimulated reconfiguration and structural isomerization of origami dimer and trimer systems. Nano Lett. 2016, 16, 6650–6655. [Google Scholar] [CrossRef] [PubMed]
- Ozkan, A.D.; Tekinay, A.B.; Guler, M.O.; Tekin, E.D. Effects of temperature, pH and counterions on the stability of peptide amphiphile nanofiber structures. RSC Adv. 2016, 6, 104201–104214. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.; Tang, T.; Takasu, A.; Higuchi, M. pH- and thermo-induced morphological changes of an amphiphilic peptide-grafted copolymer in solution. Polym. J. 2014, 46, 52–58. [Google Scholar] [CrossRef]
- Castelletto, V.; Cheng, G.; Stain, C.; Connon, C.J.; Hamley, I.W. Self-assembly of a peptide amphiphile containing l-carnosine and its mixtures with a multilamellar vesicle forming lipid. Langmuir 2012, 28, 11599–11608. [Google Scholar] [CrossRef] [PubMed]
- Hamley, I.W.; Dehsorkhi, A.; Castelletto, V.; Furzeland, S.; Atkins, D.; Seitsonen, J.; Ruokolainen, J. Reversible helical unwinding transition of a self-assembling peptide amphiphile. Soft Matter 2013, 9, 9290–9293. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.D.; Bonde, J.S.; Ye, L. Temperature and pH controlled self-assembly of a protein-polymer biohybrid. Macromol. Chem. Phys. 2018, 219, 1700597. [Google Scholar]
- Huang, Y.J.; Mai, Y.Y.; Yang, X.W.; Beser, U.; Liu, J.Z.; Zhang, F.; Yan, D.Y.; Mullen, K.; Feng, X.L. Temperature-dependent multidimensional self-assembly of polyphenylene-based “rod-coil” graft polymers. J. Am. Chem. Soc. 2015, 137, 11602–11605. [Google Scholar] [CrossRef]
- Zhong, J.; Liu, X.W.; Wei, D.X.; Yan, J.; Wang, P.; Sun, G.; He, D.N. Effect of incubation temperature on the self-assembly of regenerated silk fibroin: A study using afm. Int. J. Biol. Macromol. 2015, 76, 195–202. [Google Scholar] [CrossRef]
- Putri, R.M.; Cornelissen, J.J.L.M.; Koay, M.S.T. Self-assembled cage-like protein structures. ChemPhysChem 2015, 16, 911–918. [Google Scholar] [CrossRef]
- Semerdzhiev, S.A.; Dekker, D.R.; Subramaniam, V.; Claessens, M.M.A.E. Self-assembly of protein fibrils into suprafibrillar aggregates: Bridging the nano- and mesoscale. ACS Nano 2014, 8, 5543–5551. [Google Scholar] [CrossRef]
- Dai, B.; Li, D.; Xi, W.; Luo, F.; Zhang, X.; Zou, M.; Cao, M.; Hu, J.; Wang, W.Y.; Wei, G.H.; et al. Tunable assembly of amyloid-forming peptides into nanosheets as a retrovirus carrier. Proc. Natl. Acad. Sci. USA 2015, 112, 2996–3001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.F.; Li, Y.L.; Wang, Y.; Zheng, J.W.; Mao, C.D. Regulating DNA self-assembly by DNA-surface interactions. ChemBioChem 2017, 18, 2404–2407. [Google Scholar] [CrossRef] [PubMed]
- Garmann, R.F.; Comas-Garcia, M.; Gopal, A.; Knobler, C.M.; Gelbart, W.M. The assembly pathway of an icosahedral single-stranded rna virus depends on the strength of inter-subunit attractions. J. Mol. Biol. 2014, 426, 1050–1060. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Liu, W.Y.; Nixon, R.; Wang, R.S. Metal-ion responsive reversible assembly of DNA origami dimers: G-quadruplex induced intermolecular interaction. Nanoscale 2018, 10, 3626–3630. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.H.; Cui, Y.; He, Q.; Wang, K.W.; Li, J.B. Organogels based on self-assembly of diphenylalanine peptide and their application to immobilize quantum dots. Chem. Mater. 2008, 20, 1522–1526. [Google Scholar] [CrossRef]
- Zhu, P.L.; Yan, X.H.; Su, Y.; Yang, Y.; Li, J.B. Solvent-induced structural transition of self-assembled dipeptide: From organogels to microcrystals. Chem. Eur. J. 2010, 16, 3176–3183. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.L.; Qi, W.; Su, R.X.; Zhao, J.; He, Z.M. Solvent and surface controlled self-assembly of diphenylalanine peptide: From microtubes to nanofibers. Soft Matter 2011, 7, 6418–6421. [Google Scholar] [CrossRef]
- Mason, T.O.; Chirgadze, D.Y.; Levin, A.; Adler-Abramovich, L.; Gazit, E.; Knowles, T.P.J.; Buell, A.K. Expanding the solvent chemical space for self-assembly of dipeptide nanostructures. ACS Nano 2014, 8, 1243–1253. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Yan, X.H.; Wang, A.H.; Fei, J.B.; Cui, Y.; He, Q.; Li, J.B. A peony-flower-like hierarchical mesocrystal formed by diphenylalanine. J. Mater. Chem. 2010, 20, 6734–6740. [Google Scholar] [CrossRef]
- Ryu, J.; Park, C.B. High-temperature self-assembly of peptides into vertically well-aligned nanowires by aniline vapor. Adv. Mater. 2008, 20, 3754–3758. [Google Scholar] [CrossRef]
- Helbing, C.; Deckert-Gaudig, T.; Firkowska-Boden, I.; Wei, G.; Deckert, V.; Jandt, K.D. Protein handshake on the nanoscale: How albumin and hemoglobin self-assemble into nanohybrid fibers. ACS Nano 2018, 12, 1211–1219. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, K.; Yan, L.Y.; Wang, A.H.; Bai, S.; Yan, X.H. Trace solvent as a predominant factor to tune dipeptide self-assembly. ACS Nano 2016, 10, 2138–2143. [Google Scholar] [CrossRef] [PubMed]
- Fu, I.W.; Markegard, C.B.; Nguyen, H.D. Solvent effects on kinetic mechanisms of self-assembly by peptide amphiphiles via molecular dynamics simulations. Langmuir 2015, 31, 315–324. [Google Scholar] [CrossRef] [PubMed]
- He, H.J.; Xu, B. Instructed-assembly (IA): A molecular process for controlling cell fate. Bull. Chem. Soc. Jpn. 2018, 91, 900–906. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.E.; Gianneschi, N.C. Enzyme-directed assembly and manipulation of organic nanomaterials. Chem. Commun. 2011, 47, 11814–11821. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.M.; Gu, H.W.; Fu, D.G.; Gao, P.; Lam, J.K.; Xu, B. Enzymatic formation of supramolecular hydrogels. Adv. Mater. 2004, 16, 1440–1444. [Google Scholar] [CrossRef]
- Amir, R.J.; Zhong, S.; Pochan, D.J.; Hawker, C.J. Enzymatically triggered self-assembly of block copolymers. J. Am. Chem. Soc. 2009, 131, 13949–13951. [Google Scholar] [CrossRef]
- Guilbaud, J.B.; Vey, E.; Boothroyd, S.; Smith, A.M.; Ulijn, R.V.; Saiani, A.; Miller, A.F. Enzymatic catalyzed synthesis and triggered gelation of ionic peptides. Langmuir 2010, 26, 11297–11303. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.X.; Zhou, Z.; Wu, B.; He, B.F. Enzymatic formation of a novel cell-adhesive hydrogel based on small peptides with a laterally grafted l-3,4-dihydroxyphenylalanine group. Nanoscale 2014, 6, 1277–1280. [Google Scholar] [CrossRef] [PubMed]
- Chien, M.P.; Rush, A.M.; Thompson, M.P.; Gianneschi, N.C. Programmable shape-shifting micelles. Angew. Chem. Int. Ed. 2010, 49, 5076–5080. [Google Scholar] [CrossRef] [PubMed]
- Heck, T.; Faccio, G.; Richter, M.; Thony-Meyer, L. Enzyme-catalyzed protein crosslinking. Appl. Microbiol. Biotechnol. 2013, 97, 461–475. [Google Scholar] [CrossRef]
- Yuan, D.; Shi, J.F.; Du, X.W.; Huang, Y.B.; Gao, Y.; Baoum, A.A.; Xu, B. The enzyme-instructed assembly of the core of yeast prion sup35 to form supramolecular hydrogels. J. Mater. Chem. B 2016, 4, 1318–1323. [Google Scholar] [CrossRef] [PubMed]
- He, H.J.; Wang, H.M.; Zhou, N.; Yang, D.S.; Xu, B. Branched peptides for enzymatic supramolecular hydrogelation. Chem. Commun. 2018, 54, 86–89. [Google Scholar] [CrossRef]
- He, H.J.; Wang, J.Q.; Wang, H.M.; Zhou, N.; Yang, D.; Green, D.R.; Xu, B. Enzymatic cleavage of branched peptides for targeting mitochondria. J. Am. Chem. Soc. 2018, 140, 1215–1218. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Du, X.W.; Chen, X.Y.; Wang, J.Q.; Zhou, N.; Wu, D.F.; Xu, B. Enzymatic self-assembly confers exceptionally strong synergism with nf-kappa b targeting for selective necroptosis of cancer cells. J. Am. Chem. Soc. 2018, 140, 2301–2308. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.L.; Yan, Y.F.; Cheng, B.C.; Deng, L.F.; Shao, Z.W.; Sun, Z.L.; Li, X.M. Enzymatic formation of an injectable hydrogel from a glycopeptide as a biomimetic scaffold for vascularization. ACS Appl. Mater. Interfaces 2018, 10, 6180–6189. [Google Scholar] [CrossRef] [PubMed]
- Muraoka, T.; Cui, H.; Stupp, S.I. Quadruple helix formation of a photoresponsive peptide amphiphile and its light-triggered dissociation into single fibers. J. Am. Chem. Soc. 2008, 130, 2946–2947. [Google Scholar] [CrossRef]
- Ma, H.C.; Fei, J.B.; Li, Q.; Li, J.B. Photo-induced reversible structural transition of cationic diphenylalanine peptide self-assembly. Small 2015, 11, 1787–1791. [Google Scholar] [CrossRef]
- Tanaka, F.; Mochizuki, T.; Liang, X.G.; Asanuma, H.; Tanaka, S.; Suzuki, K.; Kitamura, S.; Nishikawa, A.; Ui-Tei, K.; Hagiya, M. Robust and photocontrollable DNA capsules using azobenzenes. Nano Lett. 2010, 10, 3560–3565. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.Y.; Endo, M.; Hidaka, K.; Sugiyama, H. Photo-controllable DNA origami nanostructures assembling into predesigned multiorientational patterns. J. Am. Chem. Soc. 2012, 134, 20645–20653. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Endo, M.; Yang, Y.Y.; Sugiyama, H. Dynamic assembly/disassembly processes of photoresponsive DNA origami nanostructures directly visualized on a lipid membrane surface. J. Am. Chem. Soc. 2014, 136, 1714–1717. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.Q.; Zhang, Y.N.; Tian, L.L.; Zhao, Y.Y.; Wu, D.N.; Xue, W.; Ramakrishna, S.; Wu, W.T.; He, L.M. Self-assembly behaviors of molecular designer functional rada16-i peptides: Influence of motifs, pH, and assembly time. Biomed. Mater. 2017, 12, 015007. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Jia, H.Y.; Cao, T.Y.; Liu, D.S. Supramolecular hydrogels based on DNA self-assembly. Acc. Chem. Res. 2017, 50, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Zhang, Z.F.; Li, D.P.; Zhang, W.S.; Yu, X.Q.; Liu, W.; Gong, C.C.; Wei, G.; Su, Z.Q. Biomimetic Ultralight, Highly Porous, Shape-Adjustable, and Biocompatible 3D Graphene Minerals via Incorporation of Self-Assembled Peptide Nanosheets. Adv. Funct. Mater. 2018, 28, 1801056. [Google Scholar] [CrossRef]
- Hamley, I.W. Small bioactive peptides for biomaterials design and therapeutics. Chem. Rev. 2017, 117, 14015–14041. [Google Scholar] [CrossRef]
- Gong, C.C.; Sun, S.W.; Zhang, Y.J.; Sun, L.; Su, Z.Q.; Wu, A.G.; Wei, G. Hierarchical nanomaterials via biomolecular self-assembly and bioinspiration for energy and environmental applications. Nanocale 2019, 11. [Google Scholar] [CrossRef]












| Biomolecules | Nanostructures | Interactions | Stimulations | Ref. |
|---|---|---|---|---|
| Proteins | ||||
| SP1 | Nanowires | Electrostatic | Micelles | [60] |
| SP1 | Nanowire-QDs | Electrostatic | Enzyme | [61] |
| BSA | NPs | Hydrophobic | Organic | [72] |
| IgG | 2D crystals | Ligand–receptor | - | [99] |
| RIDC3 | Nanotubes/2D Crystals | Zn2+-coordination | pH | [117,118] |
| Amelogenin | Nanospheres | - | pH and temperature | [126] |
| Silk fibroin | Protofibrils/Fibers | - | temperature | [128] |
| A-synuclein | Fibrils | Electrostatic | Ions | [130] |
| Peptides | ||||
| FF | Fibers/Tubes/Rods | Hydrogen bonds | Organic | [21] |
| FF | PNWs-G | Hydrogen bonds and π−π interaction | Organic | [52] |
| VIAGASLWWSEKLVIA | GN-PNF-AgNW | Electrostatic | Ethanol | [58] |
| NapFFKYp | Nanofibers | Hydrophobic | Organic | [65] |
| EAK 16-II | Nanofibers | Electrostatic/hydrophobic | Molecular structure | [66] |
| RGDAEAKAEAKYWYAFAEAKAEAKRGD | PNF-GQDs | π–π/Electrostatic | ethanol | [76] |
| AEAKAEAKYWYAFAEAKAEAK | GO-PNF | π–π/Electrostatic | Ethanol | [33] |
| AEAKAEAKYWYAFAEAKAEAK | GQD-PNF-GO | π–π/Electrostatic | Ethanol | [77] |
| Peptide | Fibers/Aggregates | Ligand–receptor | Enzyme | [96] |
| KLVFFAE | Nanofibers/Tubes | Electrostatic | pH | [112] |
| PA | Micelles/Nanofibers | Electrostatic | pH | [114] |
| C16-KKFFVLK | Nanotubes/Helical ribbons | Hydrogen bonds | Temperature | [125] |
| KLVFFAK | Nanosheets | Electrostatic | Ionic strength | [131] |
| GNNQQNY | Hydrogels | Hydrogen bonds | Enzyme | [152] |
| FFDY(H2PO3) | Fibers/Hydrogels | π–π/Hydrogen bonds | Enzyme | [156] |
| GV3A3E3 | Fibers | Hydrogen bonds/hydrophobic | Light | [157] |
| FF | Nanoplates/belts | Hydrogen bonds/π–π | Light | [158] |
| DNA/RNA | ||||
| DNA | GQDs-ionic liquid (IL)-NF-DNA | π–π interactions | Enzyme | [81] |
| DNA | GO-DNA | π–π interactions | Temperature | [82] |
| DNA | Hydrogels | Clamped hybridization | - | [24] |
| DNA | 2D lattices | base pairing | Buffer/Mg2+ | [83] |
| DNA | Tiles | base pairing | Mg2+ | [84] |
| DNA | Nanowires/Sheets | base pairing | - | [85] |
| DNA | 2D arrays | base pairing | Ni2+ | [132] |
| DNA | Capsules | base pairing | Light | [159] |
| DNA | Origami | base pairing | Light | [160] |
| DNA | Origami | base pairing | Light | [161] |
| RNA | Tetrahedrons | RNA packing | - | [91] |
| RNA | Triangles | RNA packing | - | [92] |
| RNA | Lattices/Tubes | RNA packing | - | [93] |
| PNA | Fibers | π–π and base pairing | - | [94] |
| Virus | ||||
| CCMV | 3D crystals | Ligand-receptor | - | [95] |
| Bacteriophage P22 | P22VLP-NPs | Electrostatic interaction | NPs | [59] |
| Enzymes | ||||
| OxOx/HRP | CRGO-enzyme | Hydrophobic | pH | [73] |
| GOx/CAT | graphene nanodots-porous gold | π-π | Organic | [80] |
| Other biopolymers | ||||
| cholesterol | Microrods/ribbons | - | Polymer | [104] |
| cholesterol | Aggregates | - | Polymer | [105] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Gong, C.; Yuan, X.; Wei, G. Controlling the Self-Assembly of Biomolecules into Functional Nanomaterials through Internal Interactions and External Stimulations: A Review. Nanomaterials 2019, 9, 285. https://doi.org/10.3390/nano9020285
Wang L, Gong C, Yuan X, Wei G. Controlling the Self-Assembly of Biomolecules into Functional Nanomaterials through Internal Interactions and External Stimulations: A Review. Nanomaterials. 2019; 9(2):285. https://doi.org/10.3390/nano9020285
Chicago/Turabian StyleWang, Li, Coucong Gong, Xinzhu Yuan, and Gang Wei. 2019. "Controlling the Self-Assembly of Biomolecules into Functional Nanomaterials through Internal Interactions and External Stimulations: A Review" Nanomaterials 9, no. 2: 285. https://doi.org/10.3390/nano9020285
APA StyleWang, L., Gong, C., Yuan, X., & Wei, G. (2019). Controlling the Self-Assembly of Biomolecules into Functional Nanomaterials through Internal Interactions and External Stimulations: A Review. Nanomaterials, 9(2), 285. https://doi.org/10.3390/nano9020285