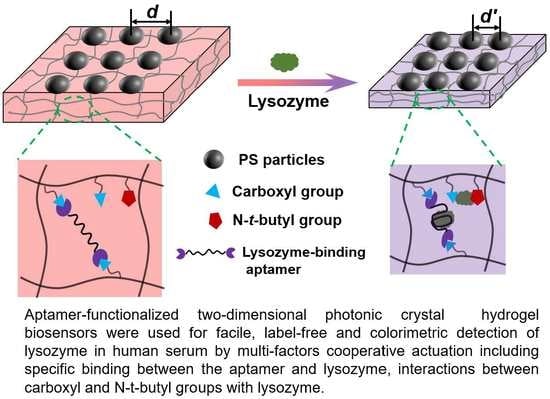
Multi-Factors Cooperatively Actuated Photonic Hydrogel Aptasensors for Facile, Label-Free and Colorimetric Detection of Lysozyme
Abstract
:1. Introduction
2. Experimental Methods
2.1. Materials and Characterization
2.2. Preparation of the Photonic Hydrogel Aptasensors
2.3. Measurement of Debye Diffraction Ring
3. Results and Discussion
3.1. Preparation and Characterization of the Photonic Hydrogel Aptasensors
3.2. Response Mechanism of the Aptasensor
3.3. Response Performance of DNA−H1−960 toward Lysozyme
3.4. Detection of Lysozyme in Human Serum
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cai, Z.; Smith, N.L.; Zhang, J.-T.; Asher, S.A. Two-Dimensional Photonic Crystal Chemical and Biomolecular Sensors. Anal. Chem. 2015, 87, 5013–5025. [Google Scholar] [CrossRef] [PubMed]
- Shen, P.; Zhang, Y.; Cai, Z.; Liu, R.; Xu, X.; Li, R.; Wang, J.-J.; Yang, D. Three-Dimensional/Two-Dimensional Photonic Crystal Hydrogels for Biosensing. J. Mater. Chem. C 2021, 9, 5840–5857. [Google Scholar] [CrossRef]
- Cai, Z.; Zhang, J.-T.; Xue, F.; Hong, Z.; Punihaole, D.; Asher, S.A. 2D Photonic Crystal Protein Hydrogel Coulometer for Sensing Serum Albumin Ligand Binding. Anal. Chem. 2014, 86, 4840–4847. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.; Horne, W.S.; Asher, S.A. Human Serum Phenylpyruvate Quantification Using Responsive 2D Photonic Crystal Hydrogels via Chemoselective Oxime Ligation: Progress toward Developing Phenylalanine-Sensing Elements. ACS Appl. Mater. Interfaces 2020, 12, 39612–39619. [Google Scholar] [CrossRef] [PubMed]
- Xue, F.; Meng, Z.; Wang, F.; Wang, Q.; Xue, M.; Xu, Z. A 2-D Photonic Crystal Hydrogel for Selective Sensing of Glucose. J. Mater. Chem. A 2014, 2, 9559–9565. [Google Scholar] [CrossRef]
- Yan, Z.; Xue, M.; He, Q.; Lu, W.; Meng, Z.; Yan, D.; Qiu, L.; Zhou, L.; Yu, Y. A Non-Enzymatic Urine Glucose Sensor with 2-D Photonic Crystal Hydrogel. Anal. Bioanal. Chem. 2016, 408, 8317–8323. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xiang, J.; Men, D.; Zhang, H. 2D Au Nanosphere Arrays/PVA-PBA-Modified-Hydrogel Composite Film for Glucose Detection with Strong Diffraction Intensity and Linear Response. Nanomaterials 2019, 9, 140. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.-T.; Cai, Z.; Kwak, D.H.; Liu, X.; Asher, S.A. Two-Dimensional Photonic Crystal Sensors for Visual Detection of Lectin Concanavalin A. Anal. Chem. 2014, 86, 9036–9041. [Google Scholar] [CrossRef]
- Cai, Z.; Sasmal, A.; Liu, X.; Asher, S.A. Responsive Photonic Crystal Carbohydrate Hydrogel Sensor Materials for Selective and Sensitive Lectin Protein Detection. ACS Sens. 2017, 2, 1474–1481. [Google Scholar] [CrossRef]
- Li, G.; Xiao, F.; Liao, S.; Chen, Q.; Zhou, J.; Wu, Z.; Yu, R. Label-Free 2D Colloidal Photonic Crystal Hydrogel Biosensor for Urea and Urease Inhibitor. Sens. Actuators B Chem. 2018, 277, 591–597. [Google Scholar] [CrossRef]
- Cai, Z.; Kwak, D.H.; Punihaole, D.; Hong, Z.; Velankar, S.S.; Liu, X.; Asher, S.A. A Photonic Crystal Protein Hydrogel Sensor for Candida albicans. Angew. Chem. Int. Ed. 2015, 54, 13036–13040. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, G.; Rizvi, A.S.; Irfan, M.; Yan, D.; Khan, R.U.; Rafique, B.; Xue, M.; Meng, Z.H.; Qu, F. Glycated Albumin Based Photonic Crystal Sensors for Detection of Lipopolysaccharides and Discrimination of Gram-Negative Bacteria. Anal. Chim. Acta 2020, 1117, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Cai, Z.; Zhang, Q.; Yuan, H.; Zhang, G.; Yang, D. Colorimetric Two-Dimensional Photonic Crystal Biosensors for Label-Free Detection of Hydrogen Peroxide. Sens. Actuators B Chem. 2022, 354, 131236. [Google Scholar] [CrossRef]
- Nimjee, S.M.; White, R.R.; Becker, R.C.; Sullenger, B.A. Aptamers as Therapeutics. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Shaban, S.M.; Kim, D.-H. Recent Advances in Aptamer Sensors. Sensors 2021, 21, 979. [Google Scholar] [CrossRef]
- Soni, S.; Jain, U.; Burke, D.H.; Chauhan, N. A Label Free, Signal off Electrochemical Aptasensor for Amphetamine Detection. Surf. Interfaces 2022, 31, 102023. [Google Scholar] [CrossRef]
- Xia, Y.; Gan, S.; Xu, Q.; Qiu, X.; Gao, P.; Huang, S. A Three-Way Junction Aptasensor for Lysozyme Detection. Biosens. Bioelectron. 2013, 39, 250–254. [Google Scholar] [CrossRef]
- Soni, S.; Jain, U.; Burke, D.H.; Chauhan, N. Development of Nanomaterial-Modified Impedimetric Aptasensor—A Single-Step Strategy for 3,4-Methylenedioxymethylamphetamine Detection. Biosensors 2022, 12, 538. [Google Scholar] [CrossRef]
- Chen, Y.; Li, H.; Gao, T.; Zhang, T.; Xu, L.; Wang, B.; Wang, J.; Pei, R. Selection of DNA Aptamers for the Development of Light-up Biosensor to Detect Pb(II). Sens. Actuators B Chem. 2018, 254, 214–221. [Google Scholar] [CrossRef]
- Rezaei, B.; Shahshahanipour, M.; Ensafi, A.A.; Farrokhpour, H. Development of Highly Selective and Sensitive Fluorimetric Label-Free Mercury Aptasensor Based on Cysteamine@CdTe/ZnS Quantum Dots, Experimental and Theoretical Investigation. Sens. Actuators B Chem. 2017, 247, 400–407. [Google Scholar] [CrossRef]
- Mao, K.; Zhang, H.; Wang, Z.; Cao, H.; Zhang, K.; Li, X.; Yang, Z. Nanomaterial-Based Aptamer Sensors for Arsenic Detection. Biosens. Bioelectron. 2020, 148, 111785. [Google Scholar] [CrossRef]
- Luan, Y.; Wang, N.; Li, C.; Guo, X.; Lu, A. Advances in the Application of Aptamer Biosensors to the Detection of Aminoglycoside Antibiotics. Antibiotics 2020, 9, 787. [Google Scholar] [CrossRef] [PubMed]
- Rabiee, N.; Ahmadi, S.; Arab, Z.; Bagherzadeh, M.; Safarkhani, M.; Nasseri, B.; Rabiee, M.; Tahriri, M.; Webster, T.J.; Tayebi, L. Aptamer Hybrid Nanocomplexes as Targeting Components for Antibiotic/Gene Delivery Systems and Diagnostics: A Review. IJN 2020, 15, 4237–4256. [Google Scholar] [CrossRef] [PubMed]
- Urbanová, V.; Jayaramulu, K.; Schneemann, A.; Kment, Š.; Fischer, R.A.; Zbořil, R. Hierarchical Porous Fluorinated Graphene Oxide@Metal–Organic Gel Composite: Label-Free Electrochemical Aptasensor for Selective Detection of Thrombin. ACS Appl. Mater. Interfaces 2018, 10, 41089–41097. [Google Scholar] [CrossRef] [PubMed]
- Melinte, G.; Selvolini, G.; Cristea, C.; Marrazza, G. Aptasensors for Lysozyme Detection: Recent Advances. Talanta 2021, 226, 122169. [Google Scholar] [CrossRef]
- Wu, L.; Wang, Y.; Xu, X.; Liu, Y.; Lin, B.; Zhang, M.; Zhang, J.; Wan, S.; Yang, C.; Tan, W. Aptamer-Based Detection of Circulating Targets for Precision Medicine. Chem. Rev. 2021, 121, 12035–12105. [Google Scholar] [CrossRef]
- Ye, B.-F.; Zhao, Y.-J.; Cheng, Y.; Li, T.-T.; Xie, Z.-Y.; Zhao, X.-W.; Gu, Z.-Z. Colorimetric Photonic Hydrogel Aptasensor for the Screening of Heavy Metal Ions. Nanoscale 2012, 4, 5998. [Google Scholar] [CrossRef]
- Wang, C.; Li, F.; Bi, Y.; Guo, W. Reversible Modulation of 2D Photonic Crystals with a Responsive Shape-Memory DNA Hydrogel Film. Adv. Mater. Interfaces 2019, 6, 1900556. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, S.; Huang, T.; Xiao, F.; Wu, Z.; Yu, R. Construction and Research of Multiple Stimuli-Responsive 2D Photonic Crystal DNA Hydrogel Sensing Platform with Double-Network Structure and Signal Self-Expression. Anal. Chem. 2022, 94, 5530–5537. [Google Scholar] [CrossRef]
- Jang, K.; Westbay, J.H.; Asher, S.A. DNA-Crosslinked 2D Photonic Crystal Hydrogels for Detection of Adenosine Actuated by an Adenosine-Binding Aptamer. ACS Sens. 2022, 7, 1648–1656. [Google Scholar] [CrossRef]
- Murtaza, G.; Rizvi, A.S.; Xue, M.; Qiu, L.; Meng, Z. Consensus Receptor-Binding Domain-Targeted Aptamer Selection and Designing of a Photonic Crystal-Decorated Aptasensor for SARS-CoV-2. Anal. Chem. 2022, 94, 7391–7399. [Google Scholar] [CrossRef] [PubMed]
- Held, J.; van Smaalen, S. The Active Site of Hen Egg-White Lysozyme: Flexibility and Chemical Bonding. Acta Crystallogr. D Biol. Crystallogr. 2014, 70, 1136–1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levinson, S.S.; Elin, R.J.; Yam, L. Light Chain Proteinuria and Lysozymuria in a Patient with Acute Monocytic Leukemia. Clin. Chem. 2002, 48, 1131–1132. [Google Scholar] [CrossRef] [PubMed]
- Shima, K.; Hirota, M.; Fukuda, M.; Tanaka, A. Determination of Urinary Lysozyme for Potential Detection of Tubular Dysfunction in Diabetic Nephropathy. Clin. Chem. 1986, 32, 1818–1822. [Google Scholar] [CrossRef]
- Fang, M.; Zhuo, K.; Chen, Y.; Zhao, Y.; Bai, G.; Wang, J. Fluorescent Probe Based on Carbon Dots/Silica/Molecularly Imprinted Polymer for Lysozyme Detection and Cell Imaging. Anal. Bioanal. Chem. 2019, 411, 5799–5807. [Google Scholar] [CrossRef]
- Currie, G.A. Serum Lysozyme as a Marker of Host Resistance. II. Patients with Malignant Melanoma, Hypernephroma or Breast Carcinoma. Br. J. Cancer 1976, 33, 593–599. [Google Scholar] [CrossRef] [Green Version]
- Harrison, J.F.; Lunt, S.; Scott, P.; Blainey, J.D. Urinary Lysozyme, Ribonuclease, and Low-Molecular-Weight Protein in Renal Disease. Lancet 1968, 291, 371–375. [Google Scholar] [CrossRef]
- Boushell, V.; Pang, S.; He, L. Aptamer-Based SERS Detection of Lysozyme on a Food-Handling Surface. J. Food Sci. 2017, 82, 225–231. [Google Scholar] [CrossRef]
- Mishra, R.K.; Hayat, A.; Mishra, G.K.; Catanante, G.; Sharma, V.; Marty, J.-L. A Novel Colorimetric Competitive Aptamer Assay for Lysozyme Detection Based on Superparamagnetic Nanobeads. Talanta 2017, 165, 436–441. [Google Scholar] [CrossRef]
- Li, X.; Zhang, H.; Zhao, Y.; Lian, L.; Wang, X.; Gao, W.; Zhu, B.; Lou, D. Design and Synthesis of Ag Nanocluster Molecular Beacon for Adenosine Triphosphate Detection. J. Anal. Methods Chem. 2019, 2019, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Sapkota, K.; Dhakal, S. FRET-Based Aptasensor for the Selective and Sensitive Detection of Lysozyme. Sensors 2020, 20, 914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirk, K.A.; Vasilescu, A.; Andreescu, D.; Senarathna, D.; Mondal, S.; Andreescu, S. Collision-Based Electrochemical Detection of Lysozyme Aggregation. Anal. Chem. 2021, 93, 2026–2037. [Google Scholar] [CrossRef] [PubMed]
- Vidal, M.-L.; Gautron, J.; Nys, Y. Development of an ELISA for Quantifying Lysozyme in Hen Egg White. J. Agric. Food Chem. 2005, 53, 2379–2385. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Sang, S.; Yuan, Z.; Duan, Q.; Guo, X.; Zhang, H.; Zhao, C. Magnetoelastic Immunosensor via Antibody Immobilization for the Specific Detection of Lysozymes. ACS Sens. 2021, 6, 3933–3939. [Google Scholar] [CrossRef]
- Zhang, F.; Cao, L.; Yang, W. Preparation of Monodisperse and Anion-Charged Polystyrene Microspheres Stabilized with Polymerizable Sodium Styrene Sulfonate by Dispersion Polymerization. Macromol. Chem. Phys. 2010, 211, 744–751. [Google Scholar] [CrossRef]
- Zhang, J.-T.; Wang, L.; Lamont, D.N.; Velankar, S.S.; Asher, S.A. Fabrication of Large-Area Two-Dimensional Colloidal Crystals. Angew. Chem. Int. Ed. 2012, 124, 6221–6224. [Google Scholar] [CrossRef]
- Mao, Y.; Daniel, L.N.; Whittaker, N.; Saffiottil, U. DNA Binding to Crystalline Silica Characterized by Fourier-Transform Infrared Spectroscopy. Environ. Health Perspect. 1994, 102, 7. [Google Scholar]
- Thomas, L.C.; Chittenden, R.A. Characteristic Infrared Absorption Frequencies of Organophosphorus Compounds—I The Phosphoryl (P=O) Group. Spectrochim. Acta A 1964, 20, 467–487. [Google Scholar] [CrossRef]
- Babić, S.D.; Serec, K. Sodium and Manganese Salt DNA Thin Films: An Infrared Spectroscopy Study. Spectrochim. Acta A 2020, 241, 118646. [Google Scholar] [CrossRef]
- Muntean, C.M.; Ştefan, R.; Tǎbǎran, A.; Tripon, C.; Bende, A.; Fǎlǎmaş, A.; Colobǎţiu, L.M.; Olar, L.E. The Influence of UV Femtosecond Laser Pulses on Bacterial DNA Structure, as Proved by Fourier Transform Infrared (FT-IR) Spectroscopy. ChemistrySelect 2021, 6, 6957–6972. [Google Scholar] [CrossRef]
- Zuo, L.; Qin, G.; Lan, Y.; Wei, Y.; Dong, C. A Turn-on Phosphorescence Aptasensor for Ultrasensitive Detection of Lysozyme in Humoral Samples. Sen. Actuators B Chem. 2019, 289, 100–105. [Google Scholar] [CrossRef]
- Wang, Z.; Meng, Z.; Xue, M.; Zhang, H.; Shea, K.J.; Kang, L. Detection of Lysozyme in Body Fluid Based on Two-Dimensional Colloidal Crystal Sensor. Microchem. J. 2020, 157, 105073. [Google Scholar] [CrossRef]
- Yoshimatsu, K.; Lesel, B.K.; Yonamine, Y.; Beierle, J.M.; Hoshino, Y.; Shea, K.J. Temperature-Responsive “Catch and Release” of Proteins by Using Multifunctional Polymer-Based Nanoparticles. Angew. Chem. Int. Ed. 2012, 124, 2455–2458. [Google Scholar] [CrossRef]
- Chen, W.; Shea, K.J.; Xue, M.; Qiu, L.; Lan, Y.; Meng, Z. Self-Assembly of the Polymer Brush-Grafted Silica Colloidal Array for Recognition of Proteins. Anal. Bioanal. Chem. 2017, 409, 5319–5326. [Google Scholar] [CrossRef] [PubMed]
- Gu, P.; Liu, X.; Tian, Y.; Zhang, L.; Huang, Y.; Su, S.; Feng, X.; Fan, Q.; Huang, W. A Novel Visible Detection Strategy for Lysozyme Based on Gold Nanoparticles and Conjugated Polymer Brush. Sens. Actuators B Chem. 2017, 246, 78–84. [Google Scholar] [CrossRef]
- Ardekani, L.S.; Moghadam, T.T.; Thulstrup, P.W.; Ranjbar, B. Design and Fabrication of a Silver Nanocluster-Based Aptasensor for Lysozyme Detection. Plasmonics 2019, 14, 1765–1774. [Google Scholar] [CrossRef]
- Xie, Y.; An, J.; Shi, P.; Ye, N. Determination of Lysozyme by Graphene Oxide–Polyethylene Glycol-Based Fluorescence Resonance Energy Transfer. Anal. Lett. 2017, 50, 148–160. [Google Scholar] [CrossRef]
- Shrivas, K.; Nirmalkar, N.; Deb, M.K.; Dewangan, K.; Nirmalkar, J.; Kumar, S. Application of Functionalized Silver Nanoparticles as a Biochemical Sensor for Selective Detection of Lysozyme Protein in Milk Sample. Spectrochim. Acta A 2019, 213, 127–133. [Google Scholar] [CrossRef]
- Chen, L.; Xia, N.; Li, T.; Bai, Y.; Chen, X. Aptasensor for Visual and Fluorometric Determination of Lysozyme Based on the Inner Filter Effect of Gold Nanoparticles on CdTe Quantum Dots. Microchim. Acta 2016, 183, 2917–2923. [Google Scholar] [CrossRef]
- Kasibabu, B.S.B.; Bhamore, J.R.; D’souza, S.L.; Kailasa, S.K. Dicoumarol Assisted Synthesis of Water Dispersible Gold Nanoparticles for Colorimetric Sensing of Cysteine and Lysozyme in Biofluids. RSC Adv. 2015, 5, 39182–39191. [Google Scholar] [CrossRef]
- Li, J.; Mu, X.; Chan, K.-C.; Ko, C.-C.; Li, M.-J. Sensitive Determination of Lysozyme by Using a Luminescent and Colorimetric Probe Based on the Aggregation of Gold Nanoparticles Induced by an Anionic Ruthenate(II) Complex. Microchim. Acta 2018, 185, 428. [Google Scholar] [CrossRef] [PubMed]
- Lou, T.; Qiang, H.; Chen, Z. Core-Shell Cu@Au Nanoparticles-Based Colorimetric Aptasensor for the Determination of Lysozyme. Talanta 2017, 163, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Hu, X.; Tan, L.; Liao, Q.; Chen, Z. Colorimetric Detection of Lysozyme Based on Its Effect on the Growth of Gold Nanoparticles Induced by the Reaction of Chloroauric Acid and Hydroxylamine. Microchim. Acta 2016, 183, 3135–3141. [Google Scholar] [CrossRef]
- Yao, X.; Ma, X.; Ding, C.; Jia, L. Colorimetric Determination of Lysozyme Based on the Aggregation of Gold Nanoparticles Controlled by a Cationic Polymer and an Aptamer. Microchim. Acta 2016, 183, 2353–2359. [Google Scholar] [CrossRef]
- Cox, J.C.; Ellington, A.D. Automated Selection of Anti-Protein Aptamers. Bioorg. Med. Chem. 2001, 9, 2525–2531. [Google Scholar] [CrossRef]









| Sample # | Total (g) | AAm (g) | AAc (g) | Bis (g) | TBAm (g) | H2O (g) |
|---|---|---|---|---|---|---|
| H1 | 15 | 1.20 | 0.45 | 0.045 | 0.16 | 13.15 |
| H2 | -- | 13.31 |
| Sample | Added (nM) | Found (nM) | Recovery % | RSD % (n = 3) |
|---|---|---|---|---|
| Human serum | 20 | 21.37 | 106.90 | 0.64 |
| 50 | 49.97 | 99.94 | 0.76 | |
| 80 | 82.96 | 103.70 | 1.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, P.; Shi, Y.; Li, R.; Han, B.; Ma, H.; Hou, X.; Zhang, Y.; Jiang, L. Multi-Factors Cooperatively Actuated Photonic Hydrogel Aptasensors for Facile, Label-Free and Colorimetric Detection of Lysozyme. Biosensors 2022, 12, 662. https://doi.org/10.3390/bios12080662
Shen P, Shi Y, Li R, Han B, Ma H, Hou X, Zhang Y, Jiang L. Multi-Factors Cooperatively Actuated Photonic Hydrogel Aptasensors for Facile, Label-Free and Colorimetric Detection of Lysozyme. Biosensors. 2022; 12(8):662. https://doi.org/10.3390/bios12080662
Chicago/Turabian StyleShen, Peiyan, Yuqing Shi, Ran Li, Bo Han, Haojie Ma, Xueyan Hou, Yuqi Zhang, and Lei Jiang. 2022. "Multi-Factors Cooperatively Actuated Photonic Hydrogel Aptasensors for Facile, Label-Free and Colorimetric Detection of Lysozyme" Biosensors 12, no. 8: 662. https://doi.org/10.3390/bios12080662
APA StyleShen, P., Shi, Y., Li, R., Han, B., Ma, H., Hou, X., Zhang, Y., & Jiang, L. (2022). Multi-Factors Cooperatively Actuated Photonic Hydrogel Aptasensors for Facile, Label-Free and Colorimetric Detection of Lysozyme. Biosensors, 12(8), 662. https://doi.org/10.3390/bios12080662