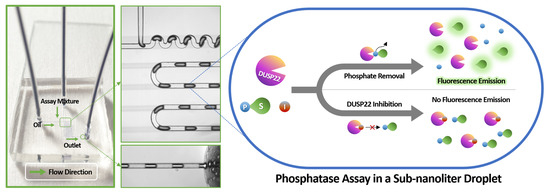
Analysis of Phosphatase Activity in a Droplet-Based Microfluidic Chip
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Purification of DUSP22 Protein
2.3. Fabrication of Microfluidic Devices and Operation
2.4. Droplet-Based Microfluidic Fluorescence Measurements
2.5. Microplate-Based Fluorescence Measurements
3. Results and Discussion
3.1. Working Principle of Phosphatase Activity Analysis in a Droplet-Based Microfluidic Chip
3.2. Droplet-Based Phosphatase Activity Assay (dPAA)
3.3. Droplet-Based Phosphatase Inhibition Assay (dPIA)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cohen, P. The role of protein phosphorylation in human health and disease. Eur. J. Biochem. 2001, 268, 5001–5010. [Google Scholar] [CrossRef] [PubMed]
- Cieśla, J.; Frączyk, T.; Rode, W. Phosphorylation of basic amino acid residues in proteins: Important but easily missed. Acta Biochim. Pol. 2011, 58, 137–148. [Google Scholar] [CrossRef]
- Alonso, A.; Rojas, A.; Godzik, A.; Mustelin, T. The dual-specific protein tyrosine phosphatase family. In Protein Phosphatases; Springer: Heidelberg, Germany, 2004; pp. 333–358. [Google Scholar]
- Patterson, K.I.; Brummer, T.; O’Brien, P.M.; Daly, R.J. Dual-specificity phosphatases: Critical regulators with diverse cellular targets. Biochem. J. 2009, 418, 475–489. [Google Scholar] [CrossRef] [PubMed]
- Raman, M.; Chen, W.; Cobb, M.H. Differential regulation and properties of MAPKs. Oncogene 2007, 26, 3100–3112. [Google Scholar] [CrossRef]
- Ostman, A.; Hellberg, C.; Böhmer, F.D. Protein-tyrosine phosphatases and cancer. Nat. Rev. Cancer 2006, 6, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.X.; Zhang, Z.Y. Targeting PTPs with small molecule inhibitors in cancer treatment. Cancer Metastasis Rev. 2008, 27, 263–272. [Google Scholar] [CrossRef]
- Liu, K.; Zheng, M.; Lu, R.; Du, J.; Zhao, Q.; Li, Z.; Li, Y.; Zhang, S. The role of CDC25C in cell cycle regulation and clinical cancer therapy: A systematic review. Cancer Cell Int. 2020, 20, 213. [Google Scholar] [CrossRef] [PubMed]
- Asante-Appiah, E.; Kennedy, B.P. Protein tyrosine phosphatases: The quest for negative regulators of insulin action. Am. J. Physiol. Endocrinol. Metab. 2003, 284, E663–E670. [Google Scholar] [CrossRef]
- Zabolotny, J.M.; Bence-Hanulec, K.K.; Stricker-Krongrad, A.; Haj, F.; Wang, Y.; Minokoshi, Y.; Kim, Y.B.; Elmquist, J.K.; Tartaglia, L.A.; Kahn, B.B.; et al. PTP1B regulates leptin signal transduction in vivo. Dev. Cell 2002, 2, 489–495. [Google Scholar] [CrossRef]
- Rodríguez-Ubreva, F.J.; Cariaga-Martinez, A.E.; Cortés, M.A.; Pablos, M.R.D.; Ropero, S.; López-Ruiz, P.; Colás, B. Knockdown of protein tyrosine phosphatase SHP-1 inhibits G1/S progression in prostate cancer cells through the regulation of components of the cell-cycle machinery. Oncogene 2010, 29, 345–355. [Google Scholar] [CrossRef] [Green Version]
- Buffet, C.; Hecale-Perlemoine, K.; Bricaire, L.; Dumont, F.; Baudry, C.; Tissier, F.; Bertherat, J.; Cochand-Priollet, B.; Raffin-Sanson, M.L.; Cormier, F.; et al. DUSP5 and DUSP6, two ERK specific phosphatases, are markers of a higher MAPK signaling activation in BRAF mutated thyroid cancers. PLoS ONE 2017, 12, e0184861. [Google Scholar] [CrossRef]
- Geladopoulos, T.P.; Sotiroudis, T.G.; Evangelopoulos, A.E. A malachite green colorimetric assay for protein phosphatase activity. Anal. Biochem. 1991, 192, 112–116. [Google Scholar] [CrossRef]
- Luchter-Wasylewska, E. Continuous assay for acid phosphatase using phenyl phosphate. Anal. Biochem. 1996, 241, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Han, S.Y.; Kato, H.; Kato, S.; Suzuki, T.; Shibata, H.; Ishii, S.; Shiiba, K.; Matsuno, S.; Kanamaru, R.; Ishioka, C. Functional evaluation of PTEN missense mutations using in vitro phosphoinositide phosphatase assay. Cancer Res. 2000, 60, 3147–3151. [Google Scholar] [PubMed]
- Hill, H.D.; Summer, G.K.; Waters, M.D. An automated fluorometric assay for alkaline phosphatase using 3-O-methylfluorescein phosphate. Anal. Biochem. 1968, 24, 9–17. [Google Scholar] [CrossRef]
- Freire, M.M.; Mignaco, J.A.; de Carvalho-Alves, P.C.; Barrabin, H.; Scofano, H.M. 3-O-Methylfluorescein phosphate as a fluorescent substrate for plasma membrane Ca2+-ATPase. Biochim. Biophys. Acta 2002, 1553, 238–248. [Google Scholar] [CrossRef]
- Moully, E.H.; Berns, E.J.; Mrksich, M. Label-free assay of protein tyrosine phosphatase activity in single cells. Anal. Chem. 2019, 91, 13206–13212. [Google Scholar] [CrossRef]
- Huebner, A.; Sharma, S.; Srisa-Art, M.; Hollfelder, F.; Edel, J.B.; deMello, A.J. Microdroplets: A sea of applications? Lab Chip 2008, 8, 1244–1254. [Google Scholar] [CrossRef]
- Cho, S.; Kang, D.K.; Choo, J.; deMello, A.J.; Chang, S.I. Recent advances in microfluidic technologies for biochemistry and molecular biologys. BMB Rep. 2011, 44, 705–712. [Google Scholar] [CrossRef]
- Hong, J.; Edel, J.B.; deMello, A.J. Micro- and nanofluidic systems for high-throughput biological screening. Drug Discov. Today 2009, 14, 134–146. [Google Scholar] [CrossRef]
- Choi, J.W.; Kang, D.K.; Park, H.; deMello, A.J.; Chang, S.I. High-throughput analysis of protein-protein interactions in picoliter-volume droplets using fluorescence polarization. Anal. Chem. 2012, 84, 3849–3854. [Google Scholar] [CrossRef] [PubMed]
- Bardiya, N.; Choi, J.W.; Chang, S.I. Analysis of single nucleotide polymorphism in human angiogenin using droplet-based microfluidics. BioChip J. 2014, 8, 15–21. [Google Scholar] [CrossRef]
- Choi, J.W.; Vasamsetti, B.M.K.; Kim, K.W.; Seo, S.H.; Lee, D.H.; Chang, S.I.; Choo, J.; Kim, H.Y. Analysis of ribonuclease activity in sub-nanoliter droplets by label-free fluorescence measurements. Analyst 2017, 142, 2610–2616. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.W.; Vasamsetti, B.M.K.; Choo, J.; Kim, H.Y. Analysis of deoxyribonuclease activity by conjugation-free fluorescence polarisation in sub-nanolitre droplets. Analyst 2020, 145, 3222–3228. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.; Cho, S. Inhibition of dual-specificity phosphatase 26 by ethyl-3,4-dephostatin: Ethyl-3,4-dephostatin as a multiphosphatase inhibitor. Pharmazie 2016, 71, 196–200. [Google Scholar]
- Garstecki, P.; Fuerstman, M.J.; Stone, H.A.; Whitesides, G.M. Formation of droplets and bubbles in a microfluidic T-junction-scaling and mechanism of break-up. Lab Chip 2006, 6, 437–446. [Google Scholar] [CrossRef]
- Sharma, S.; Srisa-Art, M.; Scott, S.; Asthana, A.; Cass, A. Droplet-based microfluidics. Methods Mol. Biol. 2013, 949, 207–230. [Google Scholar]
- Kim, B.R.; Ha, J.; Kang, E.; Cho, S. Regulation of signal transducer and activator of transcription 3 activation by dual-specificity phosphatase 3. BMB Rep. 2020, 53, 335–340. [Google Scholar] [CrossRef]
- Cha, J.H.; Jeong, Y.; Oh, A.R.; Lee, S.B.; Hong, S.S.; Kim, K. Emerging roles of PHLPP phosphatases in metabolism. BMB Rep. 2021, 54, 451–457. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vasamsetti, B.M.K.; Kim, Y.-J.; Kang, J.H.; Choi, J.-W. Analysis of Phosphatase Activity in a Droplet-Based Microfluidic Chip. Biosensors 2022, 12, 740. https://doi.org/10.3390/bios12090740
Vasamsetti BMK, Kim Y-J, Kang JH, Choi J-W. Analysis of Phosphatase Activity in a Droplet-Based Microfluidic Chip. Biosensors. 2022; 12(9):740. https://doi.org/10.3390/bios12090740
Chicago/Turabian StyleVasamsetti, Bala Murali Krishna, Yeon-Jun Kim, Jung Hoon Kang, and Jae-Won Choi. 2022. "Analysis of Phosphatase Activity in a Droplet-Based Microfluidic Chip" Biosensors 12, no. 9: 740. https://doi.org/10.3390/bios12090740
APA StyleVasamsetti, B. M. K., Kim, Y. -J., Kang, J. H., & Choi, J. -W. (2022). Analysis of Phosphatase Activity in a Droplet-Based Microfluidic Chip. Biosensors, 12(9), 740. https://doi.org/10.3390/bios12090740