Escherichia coli Enumeration in a Capillary-Driven Microfluidic Chip with SERS
Abstract
:1. Introduction
2. Experimental Section
2.1. Chemicals
2.2. Microorganisms
2.3. Instrumentation
2.4. Synthesis of Au NRs
2.5. Synthesis of Fe3O4@Au MNPs
2.6. Surface Modification Processes of Au NRs and MNPs
2.7. Preparation of Samples for TEM Measurements
2.8. Capillary-Driven Microfluidic Chip
2.9. General Procedure for the Developed Biosensor System
2.10. SERS Measurements for E. coli Detection
3. Results and Discussions
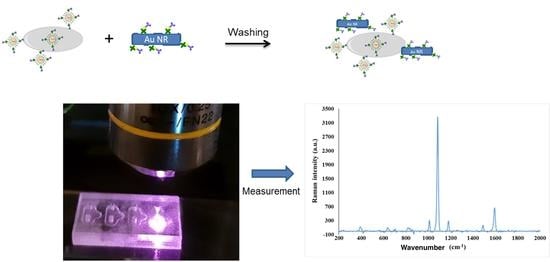
3.1. General Properties of the Chip and General Procedure for Sandwich Complex Formation
3.2. Characterizations of Au NRs and MNPs
3.3. Optimization of Analytical Parameters and Real Sample Application
3.4. Analytical Performance of the Method
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Li, Q.; Yu, S.; Han, J.; Wu, J.; You, L.; Shi, X.; Wang, S. Synergistic antibacterial activity and mechanism of action of nisin/carvacrol combination against Staphylococcus aureus and their application in the infecting pasteurized milk. Food Chem. 2022, 380, 132009. [Google Scholar] [CrossRef]
- Ruegg, P.L. The bovine milk microbiome—An evolving science. Domest. Anim. Endocrinol. 2022, 79, 106708. [Google Scholar] [CrossRef]
- Nascimento, E.C.D.; Sabino, M.C.; Corguinha, L.d.R.; Targino, B.N.; Lange, C.C.; Pinto, C.L.D.O.; Pinto, P.D.F.; Vidigal, P.M.P.; Sant’Ana, A.S.; Hungaro, H.M. Lytic bacteriophages UFJF_PfDIW6 and UFJF_PfSW6 prevent Pseudomonas fluorescens growth in vitro and the proteolytic-caused spoilage of raw milk during chilled storage. Food Microbiol. 2022, 101, 103892. [Google Scholar] [CrossRef]
- Bastam, M.M.; Jalili, M.; Pakzad, I.; Maleki, A.; Ghafourian, S. Pathogenic bacteria in cheese, raw and pasteurised milk. Vet. Med. Sci. 2021, 7, 2445–2449. [Google Scholar] [CrossRef]
- Ye, S.; Han, T.; Cheng, M.; Dong, L. Wulff-type boronic acid-functionalized quantum dots for rapid and sensitive detection of Gram-negative bacteria. Sens. Actuators B Chem. 2022, 356, 131332. [Google Scholar] [CrossRef]
- Wang, J.; Tan, L.; Bi, W.; Shen, H.; Li, D.; Yu, Z.; Gan, N. Ultrasensitive microfluidic immunosensor with stir bar enrichment for point-of-care test of Staphylococcus aureus in foods triggered by DNAzyme-assisted click reaction. Food Chem. 2022, 378, 132093. [Google Scholar] [CrossRef]
- Yan, C.; Sun, Y.; Yao, M.; Jin, X.; Yang, Q.; Wu, W. pH-responsive nanoparticles and automated detection apparatus for dual detection of pathogenic bacteria. Sens. Actuators B Chem. 2022, 354, 131117. [Google Scholar] [CrossRef]
- Zhu, A.; Ali, S.; Xu, Y.; Ouyang, Q.; Wang, Z.; Chen, Q. SERS-based Au@Ag NPs Solid-phase substrate combined with chemometrics for rapid discrimination of multiple foodborne pathogens. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 270, 120814. [Google Scholar] [CrossRef]
- Kim, S.; Ansah, I.B.; Park, J.S.; Dang, H.; Choi, N.; Lee, W.-C.; Lee, S.H.; Jung, H.S.; Kim, D.-H.; Yoo, S.M.; et al. Early and direct detection of bacterial signaling molecules through one-pot Au electrodeposition onto paper-based 3D SERS substrates. Sens. Actuators B Chem. 2022, 358, 131504. [Google Scholar] [CrossRef]
- Rho, E.; Kim, M.; Cho, S.H.; Choi, B.; Park, H.; Jang, H.; Jung, Y.S.; Jo, S. Separation-free bacterial identification in arbitrary media via deep neural network-based SERS analysis. Biosens. Bioelectron. 2022, 202, 113991. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, S.; Li, M.; Liu, W.; Wei, Z.; Zhao, L.; Liu, Y.; Xu, L.; Zhao, G.; Ma, Y. Preparation of an AgNPs@Polydimethylsiloxane (PDMS) multi-hole filter membrane chip for the rapid identification of food-borne pathogens by surface-enhanced Raman spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 267, 120456. [Google Scholar] [CrossRef]
- Jin, L.; Wang, S.; Shao, Q.; Cheng, Y. A rapid and facile analytical approach to detecting Salmonella enteritidis with aptamer-based surface-enhanced Raman spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 267, 120625. [Google Scholar] [CrossRef]
- Arslan, A.H.; Ciloglu, F.U.; Yilmaz, U.; Simsek, E.; Aydin, O. Discrimination of waterborne pathogens, Cryptosporidium parvum oocysts and bacteria using surface-enhanced Raman spectroscopy coupled with principal component analysis and hierarchical clustering. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 267, 120475. [Google Scholar] [CrossRef]
- Jia, N.; Xiong, Y.; Wang, Y.; Lu, S.; Zhang, R.; Kang, Y.; Du, Y. A novel surface-enhanced Raman scattering method for detecting fish pathogenic bacteria with Fe3O4@PEI nanocomposite and concentrated Au@Ag. J. Raman Spectrosc. 2022, 53, 211–221. [Google Scholar] [CrossRef]
- Ilhan, H.; Guven, B.; Dogan, U.; Torul, H.; Evran, S.; Çetin, D.; Suludere, Z.; Saglam, N.; Boyaci, İ.H.; Tamer, U. The coupling of immunomagnetic enrichment of bacteria with paper-based platform. Talanta 2019, 201, 245–252. [Google Scholar] [CrossRef]
- Fu, L.; Chen, Q.; Jia, L. Carbon dots and gold nanoclusters assisted construction of a ratiometric fluorescent biosensor for detection of Gram-negative bacteria. Food Chem. 2022, 374, 131750. [Google Scholar] [CrossRef]
- Kasap, E.N.; Doğan, Ü.; Çoğun, F.; Yıldırım, E.; Boyacı, İ.H.; Çetin, D.; Suludere, Z.; Tamer, U.; Ertaş, N. Fast fluorometric enumeration of E. coli using passive chip. J. Microbiol. Methods 2019, 164, 105680. [Google Scholar] [CrossRef]
- Dogan, Ü.; Kasap, E.; Cetin, D.; Suludere, Z.; Boyaci, I.H.; Türkyilmaz, C.; Ertas, N.; Tamer, U. Rapid detection of bacteria based on homogenous immunoassay using chitosan modified quantum dots. Sens. Actuators B Chem. 2016, 233, 369–378. [Google Scholar] [CrossRef]
- Dogan, Ü.; Kasap, E.N.; Sucularli, F.; Yildirim, E.; Tamer, U.; Cetin, D.; Suludere, Z.; Boyaci, I.H.; Ertas, N. Multiplex enumeration of Escherichia coli and Salmonella enteritidis in a passive capillary microfluidic chip. Anal. Methods 2020, 12, 3788–3796. [Google Scholar]
- Sun, Q.; Cheng, J.; Lin, R.; Li, J.; Zhang, Y.; Liang, X.; Su, Y.; Pang, R.; Xue, L.; Zeng, H.; et al. A novel multiplex PCR method for simultaneous identification of hypervirulent Listeria monocytogenes clonal complex 87 and CC88 strains in China. Int. J. Food Microbiol. 2022, 366, 109558. [Google Scholar]
- Arnaboldi, S.; Benevenia, R.; Bertasi, B.; Galuppini, E.; Mangeri, L.; Tilola, M.; Bassi, D.; Cocconcelli, P.S.; Stroppa, A.; Varisco, G. Validation of a real-time PCR method on pta gene for Clostridium tyrobutyricum quantification in milk. Food Control 2021, 130, 108250. [Google Scholar] [CrossRef]
- Janik, M.; Brzozowska, E.; Czyszczoń, P.; Celebańska, A.; Koba, M.; Gamian, A.; Bock, W.J.; Śmietana, M. Optical fiber aptasensor for label-free bacteria detection in small volumes. Sens. Actuators B Chem. 2021, 330, 129316. [Google Scholar] [CrossRef]
- Dursun, A.D.; Borsa, B.A.; Bayramoglu, G.; Arica, M.Y.; Ozalp, V.C. Surface plasmon resonance aptasensor for Brucella detection in milk. Talanta 2022, 239, 123074. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Ning, W.; Yang, E.; Li, Y.; Luo, Z.; Duan, Y. Sandwich method-based sensitivity enhancement of Ω-shaped fiber optic LSPR for time-flexible bacterial detection. Biosens. Bioelectron. 2022, 201, 113911. [Google Scholar] [CrossRef]
- Kumaravel, S.; Jian, S.E.; Huang, S.T.; Huang, C.H.; Hong, W.Z. Convenient and ultrasensitive detection of live Salmonella using ratiometric electrochemical molecular substrates. Anal. Chim. Acta 2022, 1190, 339244. [Google Scholar] [CrossRef]
- Jiang, H.; Jiang, D.; Liu, X.; Yang, J. A self-driven PET chip-based imprinted electrochemical sensor for the fast detection of Salmonella. Sens. Actuators B Chem. 2021, 349, 130785. [Google Scholar] [CrossRef]
- Ren, S.; Zhang, X.; Li, Z.; Jian, X.; Zhao, J.; Song, Y.Y. Development of a pulse-induced electrochemical biosensor based on gluconamide for Gram-negative bacteria detection. Microchim. Acta 2021, 188, 399. [Google Scholar] [CrossRef]
- Xu, J.; Yang, Q.; Wang, P.; Wu, D.; Yang, X.; Chen, W.; Gao, S.; Wang, S. Self-assembled β-galactosidase on T4 phage capsid through affinity binding with enhanced activity and stability for rapid bacteria detection. Sens. Actuators B Chem. 2022, 359, 131569. [Google Scholar] [CrossRef]
- Gupta, R.; Kumar, A.; Kumar, S.; Pinnaka, A.K.; Singhal, N.K. Naked eye colorimetric detection of Escherichia coli using aptamer conjugated graphene oxide enclosed Gold nanoparticles. Sens. Actuators B Chem. 2021, 329, 129100. [Google Scholar] [CrossRef]
- Tahir, M.A.; Dina, N.E.; Cheng, H.; Valev, V.K.; Zhang, L. Surface-enhanced Raman spectroscopy for bioanalysis and diagnosis. Nanoscale 2021, 13, 11593–11634. [Google Scholar] [CrossRef]
- Chen, H.; Das, A.; Bi, L.; Choi, N.; Moon, J.I.; Wu, Y.; Park, S.; Choo, J. Recent advances in surface-enhanced Raman scattering-based microdevices for point-of-care diagnosis of viruses and bacteria. Nanoscale 2020, 12, 21560–21570. [Google Scholar] [CrossRef]
- Yang, S.Z.; Liu, Q.A.; Liu, Y.L.; Weng, G.J.; Zhu, J.; Li, J.J. Recent progress in the optical detection of pathogenic bacteria based on noble metal nanoparticles. Microchim. Acta 2021, 188, 258. [Google Scholar] [CrossRef]
- El-Boubbou, K.; Gruden, C.; Huang, X. Magnetic glyco-nanoparticles: A unique tool for rapid pathogen detection, decontamination, and strain differentiation. J. Am. Chem. Soc. 2007, 129, 13392–13393. [Google Scholar] [CrossRef]
- Cheng, Y.; Liu, Y.; Huang, J.; Li, K.; Zhang, W.; Xian, Y.; Jin, L. Combining biofunctional magnetic nanoparticles and ATP bioluminescence for rapid detection of Escherichia coli. Talanta 2009, 77, 1332–1336. [Google Scholar] [CrossRef]
- Cho, Y.K.; Kim, S.; Lee, K.; Park, C.; Lee, J.G.; Ko, C. Bacteria concentration using a membrane type insulator-based dielectrophoresis in a plastic chip. Electrophoresis 2009, 30, 3153–3159. [Google Scholar] [CrossRef]
- Lin, H.Y.; Huang, C.H.; Hsieh, W.H.; Liu, L.H.; Lin, Y.C.; Chu, C.C.; Wang, S.T.; Kuo, I.T.; Chau, L.K.; Yang, C.Y. On-line SERS detection of single bacterium using novel SERS nanoprobes and a microfluidic dielectrophoresis device. Small 2014, 10, 4700–4710. [Google Scholar] [CrossRef]
- Cheng, I.F.; Chang, H.C.; Chen, T.Y.; Hu, C.; Yang, F.L. Rapid (<5 min) identification of pathogen in human blood by electrokinetic concentration and surface-enhanced Raman spectroscopy. Sci. Rep. 2013, 3, 2365. [Google Scholar]
- Krafft, B.; Tycova, A.; Urban, R.D.; Dusny, C.; Belder, D. Microfluidic device for concentration and SERS-based detection of bacteria in drinking water. Electrophoresis 2021, 42, 86–94. [Google Scholar] [CrossRef]
- Phurimsak, C.; Yildirim, E.; Tarn, M.D.; Trietsch, S.J.; Hankemeier, T.; Pamme, N.; Vulto, P. Phaseguide assisted liquid lamination for magnetic particle-based assays. Lab Chip 2014, 14, 2334–2343. [Google Scholar] [CrossRef]
- Berry, S.M.; MacCoux, L.J.; Beebe, D.J. Streamlining immunoassays with immiscible filtrations assisted by surface tension. Anal. Chem. 2012, 84, 5518–5523. [Google Scholar] [CrossRef]
- Tamer, U.; Cetin, D.; Suludere, Z.; Boyaci, I.H.; Temiz, H.T.; Yegenoglu, H.; Daniel, P.; Dinçer, I.; Elerman, Y. Gold-coated iron composite nanospheres targeted the detection of Escherichia coli. Int. J. Mol. Sci. 2013, 14, 6223–6240. [Google Scholar] [CrossRef]
- Gjergjizi, B.; Çoğun, F.; Yıldırım, E.; Eryılmaz, M.; Selbes, Y.; Sağlam, N.; Tamer, U. SERS-based ultrafast and sensitive detection of luteinizing hormone in human serum using a passive microchip. Sens. Actuators B Chem. 2018, 269, 314–321. [Google Scholar] [CrossRef]
- Ahi, E.E.; Torul, H.; Zengin, A.; Sucularlı, F.; Yıldırım, E.; Selbes, Y.; Suludere, Z.; Tamer, U. A capillary driven microfluidic chip for SERS based hCG detection. Biosens. Bioelectron. 2022, 195, 113660. [Google Scholar] [CrossRef]
- İlhan, H.; Panhwar, S.; Boyaci, İ.H.; Tamer, U. Optical Based Transducers for Biosensors. In Biosensors: Fundamentals Emerging Technologies and Applications; CRC Press: Boca Raton, FL, USA, 2022; pp. 155–186. [Google Scholar]
- Murakami, T.; Kuroda, S.I.; Osawa, Z. Dynamics of polymeric solid surfaces treated with oxygen plasma: Effect of aging media after plasma treatment. J. Colloid Interface Sci. 1998, 202, 37–44. [Google Scholar] [CrossRef]
- Fritz, J.L.; Owen, M.J. Hydrophobic recovery of plasma-treated polydimethylsiloxane. J. Adhes. 1995, 54, 33–45. [Google Scholar] [CrossRef]
- Sakamoto, H.; Hatsuda, R.; Miyamura, K.; Sugiyama, S. Plasma separation PMMA device driven by capillary force controlling surface wettability. Micro Nano Lett. 2012, 7, 64–67. [Google Scholar] [CrossRef]
- Zhang, W.; Rahmani, M.; Niu, W.; Ravaine, S.; Hong, M.; Lu, X. Tuning interior nanogaps of double-shelled Au/Ag nanoboxes for surface-enhanced Raman scattering. Sci. Rep. 2015, 5, 8382. [Google Scholar] [CrossRef]
- Brosel-Oliu, S.; Ferreira, R.; Uria, N.; Abramova, N.; Gargallo, R.; Munoz-Pascual, F.X.; Bratov, A. Novel impedimetric aptasensor for label-free detection of Escherichia coli O157:H7. Sens. Actuators B Chem. 2018, 255, 2988–2995. [Google Scholar] [CrossRef]
- Díaz-Amaya, S.; Lin, L.K.; Deering, A.J.; Stanciu, L.A. Aptamer-based SERS biosensor for whole cell analytical detection of E. coli O157:H7. Anal. Chim. Acta 2019, 1081, 146–156. [Google Scholar] [CrossRef]
- Zeinhom, M.M.A.; Wang, Y.; Song, Y.; Zhu, M.J.; Lin, Y.; Du, D. A portable smart-phone device for rapid and sensitive detection of E. coli O157:H7 in Yoghurt and Egg. Biosens. Bioelectron. 2018, 99, 479–485. [Google Scholar] [CrossRef]
- Zhou, C.; Zou, H.; Li, M.; Sun, C.; Ren, D.; Li, Y. Fiber optic surface plasmon resonance sensor for detection of E. coli O157:H7 based on antimicrobial peptides and AgNPs-rGO. Biosens. Bioelectron. 2018, 117, 347–353. [Google Scholar] [CrossRef]
- Yao, L.; Wang, L.; Huang, F.; Cai, G.; Xi, X.; Lin, J. A microfluidic impedance biosensor based on immunomagnetic separation and urease catalysis for continuous-flow detection of E. coli O157:H7. Sens. Actuators B Chem. 2018, 259, 1013–1021. [Google Scholar] [CrossRef]
- Kaushik, S.; Pandey, A.; Tiwari, U.K.; Sinha, R.K.; İlhan, H.; Panhwar, S.; Boyaci, İ.H.; Tamer, U. A label-free fiber optic biosensor for Salmonella Typhimurium detection. Opt. Fiber Technol. 2018, 46, 95–103. [Google Scholar] [CrossRef]
- Guven, B.; Basaran-Akgul, N.; Temur, E.; Tamer, U.; Boyacı, I.H. SERS-based sandwich immunoassay using antibody coated magnetic nanoparticles for Escherichia coli enumeration. Analyst 2011, 136, 740–748. [Google Scholar] [CrossRef]
- Yang, D.; Zhou, H.; Haisch, C.; Niessner, R.; Ying, Y. Reproducible, E. coli detection based on label-free SERS and mapping. Talanta 2016, 146, 457–463. [Google Scholar] [CrossRef]







| Spiked E. coli Concentration (cfu/mL) | Detected E. coli Concentration (cfu/mL) | Recovery (%) |
|---|---|---|
| 1.2 × 102 | 1.4 ± 0.3 × 102 | 113 |
| 1.2 × 104 | 1.5 ± 0.4 × 104 | 120 |
| Total Analysis Time | LOD (cfu mL−1) | Dynamic Range (cfu mL−1) | References |
|---|---|---|---|
| 30 min | 2.9 × 102 | 101–105 | [49] |
| 20 min | 101 | 102–106 | [50] |
| 2 h | 1 | 100–106 | [51] |
| Less than 1 h | 5 × 102 | 103–5 × 107 | [52] |
| 5 min | 1.2 × 101 | 101–105 | [53] |
| 20 min | 2.5 × 102 | 5 × 102–5 × 103 | [54] |
| Less than 70 min | 8 | 101–104 | [55] |
| 3 h | 105 | 105–107 | [56] |
| Less than 1 h | 7 | 101–107 | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dogan, Ü.; Sucularlı, F.; Yildirim, E.; Cetin, D.; Suludere, Z.; Boyaci, I.H.; Tamer, U. Escherichia coli Enumeration in a Capillary-Driven Microfluidic Chip with SERS. Biosensors 2022, 12, 765. https://doi.org/10.3390/bios12090765
Dogan Ü, Sucularlı F, Yildirim E, Cetin D, Suludere Z, Boyaci IH, Tamer U. Escherichia coli Enumeration in a Capillary-Driven Microfluidic Chip with SERS. Biosensors. 2022; 12(9):765. https://doi.org/10.3390/bios12090765
Chicago/Turabian StyleDogan, Üzeyir, Ferah Sucularlı, Ender Yildirim, Demet Cetin, Zekiye Suludere, Ismail Hakkı Boyaci, and Ugur Tamer. 2022. "Escherichia coli Enumeration in a Capillary-Driven Microfluidic Chip with SERS" Biosensors 12, no. 9: 765. https://doi.org/10.3390/bios12090765
APA StyleDogan, Ü., Sucularlı, F., Yildirim, E., Cetin, D., Suludere, Z., Boyaci, I. H., & Tamer, U. (2022). Escherichia coli Enumeration in a Capillary-Driven Microfluidic Chip with SERS. Biosensors, 12(9), 765. https://doi.org/10.3390/bios12090765