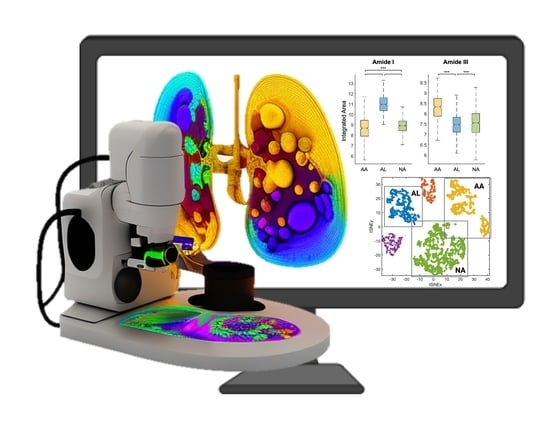
Non-Perturbative Identification and Subtyping of Amyloidosis in Human Kidney Tissue with Raman Spectroscopy and Machine Learning
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Raman Spectroscopy
2.3. Data Analysis
3. Results and Discussion
3.1. Amide I and Amide III Bands Reveal Protein Secondary Structures Associated with Amyloidosis
3.2. Machine Learning-Based Raman Spectral Analysis Can Classify Renal Amyloidosis with Respect to Deposition Sites and Types
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Merlini, G.; Bellotti, V. Molecular mechanisms of amyloidosis. N. Engl. J. Med. 2003, 349, 583–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersson, T.; Konttinen, Y.T. Amyloidosis—Recent developments. Semin. Arthritis Rheum. 2010, 39, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Dispenzieri, A.; Gertz, M.A.; Buadi, F. What do I need to know about immunoglobulin light chain (AL) amyloidosis? Blood Rev. 2012, 26, 137–154. [Google Scholar] [CrossRef] [PubMed]
- Sanchorawala, V. Light-chain (AL) amyloidosis: Diagnosis and treatment. Clin. J. Am. Soc. Nephrol. 2006, 1, 1331–1341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fändrich, M.; Nyström, S.; Nilsson, K.P.R.; Böckmann, A.; LeVine, H., III; Hammarström, P. Amyloid fibril polymorphism: A challenge for molecular imaging and therapy. J. Intern. Med. 2018, 283, 218–237. [Google Scholar] [CrossRef] [PubMed]
- Nelson, R.; Eisenberg, D. Recent atomic models of amyloid fibril structure. Curr. Opin. Struct. Biol. 2006, 16, 260–265. [Google Scholar] [CrossRef]
- Fändrich, M. On the structural definition of amyloid fibrils and other polypeptide aggregates. Cell. Mol. Life Sci. 2007, 64, 2066–2078. [Google Scholar] [CrossRef]
- Schmidt, A.; Annamalai, K.; Schmidt, M.; Grigorieff, N.; Fändrich, M. Cryo-EM reveals the steric zipper structure of a light chain-derived amyloid fibril. Proc. Natl. Acad. Sci. USA 2016, 113, 6200–6205. [Google Scholar] [CrossRef] [Green Version]
- Flynn, J.D.; McGlinchey, R.P.; Walker, R.L., 3rd; Lee, J.C. Structural features of α-synuclein amyloid fibrils revealed by Raman spectroscopy. J. Biol. Chem. 2018, 293, 767–776. [Google Scholar] [CrossRef] [Green Version]
- Chandra, B.; Korn, A.; Maity, B.K.; Adler, J.; Rawat, A.; Krueger, M.; Huster, D.; Maiti, S. Stereoisomers Probe Steric Zippers in Amyloid-β. J. Phys. Chem. B 2017, 121, 1835–1842. [Google Scholar] [CrossRef]
- Flynn, J.D.; Lee, J.C. Raman fingerprints of amyloid structures. Chem. Commun. 2018, 54, 6983–6986. [Google Scholar] [CrossRef] [PubMed]
- Kurouski, D.; Van Duyne, R.P.; Lednev, I.K. Exploring the structure and formation mechanism of amyloid fibrils by Raman spectroscopy: A review. Analyst 2015, 140, 4967–4980. [Google Scholar] [CrossRef]
- Handen, J.; Lednev, I.K. Chapter 14—Ultraviolet Raman spectroscopy for understanding structure and formation mechanism of amyloid fibrils. In Vibrational Spectroscopy in Protein Research; Ozaki, Y., Baranska, M., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 415–434. [Google Scholar]
- Devitt, G.; Howard, K.; Mudher, A.; Mahajan, S. Raman Spectroscopy: An Emerging Tool in Neurodegenerative Disease Research and Diagnosis. ACS Chem. Neurosci. 2018, 9, 404–420. [Google Scholar] [CrossRef] [PubMed]
- Gulyamov, S.; Shamshiddinova, M.; Bae, W.H.; Park, Y.C.; Kim, H.J.; Cho, W.B.; Lee, Y.M. Identification of biomarkers on kidney failure by Raman spectroscopy. J. Raman Spectrosc. 2021, 52, 1712–1721. [Google Scholar] [CrossRef]
- Sperati, C.J.; Zhang, C.; Delsante, M.; Gupta, R.; Bagnasco, S.; Barman, I. Raman spectroscopy for the diagnosis of intratubular triamterene crystallization. Kidney Int. Rep. 2018, 3, 997–1003. [Google Scholar] [CrossRef] [Green Version]
- Rygula, A.; Majzner, K.; Marzec, K.M.; Kaczor, A.; Pilarczyk, M.; Baranska, M. Raman spectroscopy of proteins: A review. J. Raman Spectrosc. 2013, 44, 1061–1076. [Google Scholar] [CrossRef]
- Talari, A.C.S.; Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2015, 50, 46–111. [Google Scholar] [CrossRef]
- Lenzi, E.; Dinarelli, S.; Longo, G.; Girasole, M.; Mussi, V. Multivariate analysis of mean Raman spectra of erythrocytes for a fast analysis of the biochemical signature of ageing. Talanta 2021, 221, 121442. [Google Scholar] [CrossRef]
- Durastanti, C.; Cirillo, E.N.M.; De Benedictis, I.; Ledda, M.; Sciortino, A.; Lisi, A.; Convertino, A.; Mussi, V. Statistical Classification for Raman Spectra of Tumoral Genomic DNA. Micromachines 2022, 13, 1388. [Google Scholar] [CrossRef]
- Devitt, G.; Rice, W.; Crisford, A.; Nandhakumar, I.; Mudher, A.; Mahajan, S. Conformational Evolution of Molecular Signatures during Amyloidogenic Protein Aggregation. ACS Chem. Neurosci. 2019, 10, 4593–4611. [Google Scholar] [CrossRef]
- Kurouski, D.; Lednev, I.K. The impact of protein disulfide bonds on the amyloid fibril morphology. Int. J. Biomed. Nanosci. Nanotechnol. 2011, 2, 167–176. [Google Scholar] [CrossRef] [Green Version]
- Ishigaki, M.; Morimoto, K.; Chatani, E.; Ozaki, Y. Exploration of Insulin Amyloid Polymorphism Using Raman Spectroscopy and Imaging. Biophys. J. 2020, 118, 2997–3007. [Google Scholar] [CrossRef] [PubMed]
- Ettema, L.; Lochocki, B.; Hoozemans, J.J.M.; de Boer, J.F.; Ariese, F. Label-free Raman and fluorescence imaging of amyloid plaques in human Alzheimer’s disease brain tissue reveal carotenoid accumulations. J. Opt. 2022, 24, 054005. [Google Scholar] [CrossRef]
- Fonseca, E.A.; Lafetá, L.; Cunha, R.; Miranda, H.; Campos, J.; Medeiros, H.G.; Romano-Silva, M.A.; Silva, R.A.; Barbosa, A.S.; Vieira, R.P.; et al. A fingerprint of amyloid plaques in a bitransgenic animal model of Alzheimer’s disease obtained by statistical unmixing analysis of hyperspectral Raman data. Analyst 2019, 144, 7049–7056. [Google Scholar] [CrossRef]
- Brauchle, E.; Sevgi, F.; Berrio, D.C.; Schenke-Layland, K.; Casadei, N.; Riess, O.; Salker, M.S.; Singh, Y. Raman imaging of α-synuclein aggregates in a rat model of Parkinson’s disease. In Proceedings of the Imaging, Manipulation, and Analysis of Biomolecules, Cells, and Tissues XVII, San Francisco, CA, USA, 4 March 2019. [Google Scholar]
- Chen, P.; Shen, A.; Zhao, W.; Baek, S.-J.; Yuan, H.; Hu, J. Raman signature from brain hippocampus could aid Alzheimer’s disease diagnosis. Appl. Opt. 2009, 48, 4743–4748. [Google Scholar] [CrossRef] [PubMed]
- Krafft, C.; Sobottka, S.B.; Schackert, G.; Salzer, R. Near infrared Raman spectroscopic mapping of native brain tissue and intracranial tumors. Analyst 2005, 130, 1070–1077. [Google Scholar] [CrossRef] [PubMed]
- Ami, D.; Lavatelli, F.; Rognoni, P.; Palladini, G.; Raimondi, S.; Giorgetti, S.; Monti, L.; Doglia, S.M.; Natalello, A.; Merlini, G. In situ characterization of protein aggregates in human tissues affected by light chain amyloidosis: A FTIR microspectroscopy study. Sci. Rep. 2016, 6, 29096. [Google Scholar] [CrossRef] [Green Version]
- Michael, R.; Lenferink, A.; Vrensen, G.; Gelpi, E.; Barraquer, R.I.; Otto, C. Hyperspectral Raman imaging of neuritic plaques and neurofibrillary tangles in brain tissue from Alzheimer’s disease patients. Sci. Rep. 2017, 7, 15603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lochocki, B.; Morrema, T.H.J.; Ariese, F.; Hoozemans, J.J.M.; de Boer, J.F. The search for a unique Raman signature of amyloid-beta plaques in human brain tissue from Alzheimer’s disease patients. Analyst 2020, 145, 1724–1736. [Google Scholar] [CrossRef] [Green Version]
- Lochocki, B.; Boon, B.D.C.; Verheul, S.R.; Zada, L.; Hoozemans, J.J.M.; Ariese, F.; de Boer, J.F. Multimodal, label-free fluorescence and Raman imaging of amyloid deposits in snap-frozen Alzheimer’s disease human brain tissue. Commun. Biol. 2021, 4, 474. [Google Scholar] [CrossRef]
- Lobanova, E.; Lobanov, S.; Triantafilou, K.; Langbein, W.; Borri, P. Quantitative chemical imaging of amyloid-β plaques with Raman micro-spectroscopy in human Alzheimer’s diseased brains. arXiv 2018, arXiv:1803.01201. [Google Scholar]
- Said, S.M.; Sethi, S.; Valeri, A.M.; Leung, N.; Cornell, L.D.; Fidler, M.E.; Herrera Hernandez, L.; Vrana, J.A.; Theis, J.D.; Quint, P.S.; et al. Renal Amyloidosis: Origin and Clinicopathologic Correlations of 474 Recent Cases. Clin. J. Am. Soc. Nephrol. 2013, 8, 1515–1523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Yu, Y.; Zhu, I.; Cheng, Y.; Sun, P.D. Structural mechanism of serum amyloid A-mediated inflammatory amyloidosis. Proc. Natl. Acad. Sci. USA 2014, 111, 5189–5194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khalighi, M.A.; Dean Wallace, W.; Palma-Diaz, M.F. Amyloid nephropathy. Clin. Kidney J. 2014, 7, 97–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sethi, S.; Theis, J.D. Pathology and diagnosis of renal non-AL amyloidosis. J. Nephrol. 2018, 31, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Beier, B.D.; Berger, A.J. Method for automated background subtraction from Raman spectra containing known contaminants. Analyst 2009, 134, 1198–1202. [Google Scholar] [CrossRef]
- Press, W.H.; Teukolsky, S.A. Savitzky-Golay smoothing filters. Comput. Phys. 1990, 4, 669–672. [Google Scholar] [CrossRef]
- Flynn, J.D.; Jiang, Z.; Lee, J.C. Segmental 13C-Labeling and Raman Microspectroscopy of α-Synuclein Amyloid Formation. Angew. Chem. Int. Ed. 2018, 57, 17069–17072. [Google Scholar] [CrossRef]
- Demšar, J.; Curk, T.; Erjavec, A.; Gorup, Č.; Hočevar, T.; Milutinovič, M.; Možina, M.; Polajnar, M.; Toplak, M.; Starič, A. Orange: Data mining toolbox in Python. J. Mach. Learn. Res. 2013, 14, 2349–2353. [Google Scholar]
- Van der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Sander, J.; Ester, M.; Kriegel, H.-P.; Xu, X. Density-based clustering in spatial databases: The algorithm gdbscan and its applications. Data Min. Knowl. Discov. 1998, 2, 169–194. [Google Scholar] [CrossRef]
- Louros, N.N.; Tsiolaki, P.L.; Baltoumas, F.A.; Chryssikos, G.D.; Gionis, V.; Hamodrakas, S.J.; Iconomidou, V.A. Tracking the amyloidogenic core of IAPP amyloid fibrils: Insights from micro-Raman spectroscopy. J. Struct. Biol. 2017, 199, 140–152. [Google Scholar] [CrossRef] [PubMed]
- Mollee, P.; Renaut, P.; Gottlieb, D.; Goodman, H. How to diagnose amyloidosis. Intern. Med. J. 2014, 44, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Holechek, M.J. Glomerular filtration: An overview. Nephrol. Nurs. J. 2003, 30, 285–290. [Google Scholar]
- Varma, V.K.; Kajdacsy-Balla, A.; Akkina, S.K.; Setty, S.; Walsh, M.J. A label-free approach by infrared spectroscopic imaging for interrogating the biochemistry of diabetic nephropathy progression. Kidney Int. 2016, 89, 1153–1159. [Google Scholar] [CrossRef] [Green Version]
- Varma, V.K.; Kajdacsy-Balla, A.; Akkina, S.; Setty, S.; Walsh, M.J. Predicting Fibrosis Progression in Renal Transplant Recipients Using Laser-Based Infrared Spectroscopic Imaging. Sci. Rep. 2018, 8, 686. [Google Scholar] [CrossRef] [Green Version]
- Guo, S.; Popp, J.; Bocklitz, T. Chemometric analysis in Raman spectroscopy from experimental design to machine learning–based modeling. Nat. Protoc. 2021, 16, 5426–5459. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.H.; Zhang, C.; Sperati, C.J.; Bagnasco, S.M.; Barman, I. Non-Perturbative Identification and Subtyping of Amyloidosis in Human Kidney Tissue with Raman Spectroscopy and Machine Learning. Biosensors 2023, 13, 466. https://doi.org/10.3390/bios13040466
Kim JH, Zhang C, Sperati CJ, Bagnasco SM, Barman I. Non-Perturbative Identification and Subtyping of Amyloidosis in Human Kidney Tissue with Raman Spectroscopy and Machine Learning. Biosensors. 2023; 13(4):466. https://doi.org/10.3390/bios13040466
Chicago/Turabian StyleKim, Jeong Hee, Chi Zhang, Christopher John Sperati, Serena M. Bagnasco, and Ishan Barman. 2023. "Non-Perturbative Identification and Subtyping of Amyloidosis in Human Kidney Tissue with Raman Spectroscopy and Machine Learning" Biosensors 13, no. 4: 466. https://doi.org/10.3390/bios13040466
APA StyleKim, J. H., Zhang, C., Sperati, C. J., Bagnasco, S. M., & Barman, I. (2023). Non-Perturbative Identification and Subtyping of Amyloidosis in Human Kidney Tissue with Raman Spectroscopy and Machine Learning. Biosensors, 13(4), 466. https://doi.org/10.3390/bios13040466