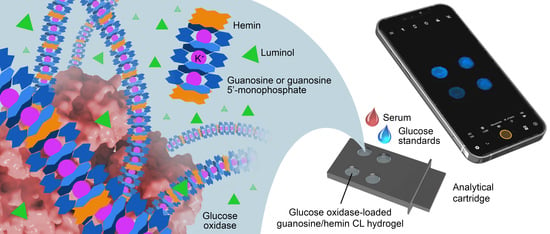
Smartphone-Based Chemiluminescence Glucose Biosensor Employing a Peroxidase-Mimicking, Guanosine-Based Self-Assembled Hydrogel
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Preparation of the Guanosine/Hemin CL Hydrogel
2.3. Measurement of H2O2 and Glucose in the Microtiter Plate Format
2.4. Smartphone-Based CL Detection with 3D Printed Device
2.5. Real Sample Analysis
2.6. Data Analysis
3. Results and Discussion
3.1. Design of the G-Quadruplex Hydrogel-Based Biosensor
3.2. G-Quadruplex Hydrogel Performance for H2O2 Quantitative Detection
3.3. G-Quadruplex Hydrogel Performance for Glucose Quantitative Detection
3.4. Smartphone-Based Glucose Biosensing Device
3.5. Assay Selectivity
3.6. Application to Real Samples
3.7. Stability of the Glucose Biosensor
3.8. Hydrogel 3D Printing
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boselli, L.; Pomili, T.; Donati, P.; Pompa, P.P. Nanosensors for visual detection of glucose in biofluids: Are we ready for instrument-free home-testing? Materials 2021, 14, 1978. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.H.; Yao, Y.; Chen, T.Y.; Shen, W.; Tang, S.; Lee, H.K. Application of smartphone-based spectroscopy to biosample analysis: A review. Biosens. Bioelectron. 2021, 172, 112788. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Neuta, I.; Neumann, F.; Brightmeyer, J.; Tis, T.B.; Madaboosi, N.; Wei, Q.; Ozcan, A.; Nilsson, M. Smartphone-based clinical diagnostics: Towards democratization of evidence-based health care. J. Intern. Med. 2019, 285, 19–39. [Google Scholar] [CrossRef] [Green Version]
- Pravda, J. Hydrogen peroxide and disease: Towards a unified system of pathogenesis and therapeutics. Mol. Med. 2020, 26, 41. [Google Scholar] [CrossRef] [PubMed]
- Lennicke, C.; Rahn, J.; Lichtenfels, R.; Wessjohann, L.A.; Seliger, B. Hydrogen peroxide—Production, fate and role in redox signaling of tumor cells. Cell Commun. Signal 2015, 13, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giorgio, M.; Trinei, M.; Migliaccio, E.; Pelicci, P.G. Hydrogen peroxide: A metabolic by-product or a common mediator of ageing signals? Nat. Rev. Mol. Cell Biol. 2007, 8, 722–728. [Google Scholar] [CrossRef]
- Miller, E.W.; Dickinson, B.C.; Chang, C.J. Aquaporin-3 mediates hydrogen peroxide uptake to regulate downstream intracellular signaling. Proc. Natl. Acad. Sci. USA 2010, 107, 15681–15686. [Google Scholar] [CrossRef] [Green Version]
- Sies, H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: Oxidative eustress. Redox Biol. 2017, 11, 613–619. [Google Scholar] [CrossRef]
- Pi, J.; Bai, Y.; Zhang, Q.; Wong, V.; Floering, L.M.; Daniel, K.; Reece, J.M.; Deeney, J.T.; Andersen, M.E.; Corkey, B.E.; et al. Reactive oxygen species as a signal in glucose-stimulated insulin secretion. Diabetes 2007, 56, 1783–1791. [Google Scholar] [CrossRef] [Green Version]
- Liou, G.Y.; Storz, P. Reactive oxygen species in cancer. Free Radic. Res. 2010, 44, 479–496. [Google Scholar] [CrossRef] [Green Version]
- Barnham, K.J.; Masters, C.L.; Bush, A.I. Neurodegenerative diseases and oxidatives stress. Nat. Rev. Drug Discov. 2004, 3, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Patel, V.; Kruse, P.; Selvaganapathy, P.R. Solid state sensors for hydrogen peroxide detection. Biosensors 2020, 11, 9. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Yuan, R.; Chai, Y.; Hu, F. Electrochemical sensing of hydrogen peroxide using metal nanoparticles: A review. Microchim. Acta 2013, 180, 15–32. [Google Scholar] [CrossRef]
- Meier, J.; Hofferber, E.M.; Stapleton, J.A.; Iverson, N.M. Hydrogen peroxide sensors for biomedical applications. Chemosensors 2019, 7, 64. [Google Scholar] [CrossRef] [Green Version]
- Roda, A.; Arduini, F.; Mirasoli, M.; Zangheri, M.; Fabiani, L.; Colozza, N.; Marchegiani, E.; Simoni, P.; Moscone, D. A challenge in biosensors: Is it better to measure a photon or an electron for ultrasensitive detection? Biosens. Bioelectron. 2020, 155, 112093. [Google Scholar] [CrossRef]
- Calabria, D.; Guardigli, M.; Mirasoli, M.; Punzo, A.; Porru, E.; Zangheri, M.; Simoni, P.; Pagnotta, E.; Ugolini, L.; Lazzeri, L.; et al. Selective chemiluminescent TURN-ON quantitative bioassay and imaging of intracellular hydrogen peroxide in human living cells. Anal. Biochem. 2020, 600, 113760. [Google Scholar] [CrossRef] [PubMed]
- Calabria, D.; Zangheri, M.; Trozzi, I.; Lazzarini, E.; Pace, A.; Mirasoli, M.; Guardigli, M. Smartphone-based chemiluminescent origami mu PAD for the rapid assessment of glucose blood levels. Biosensors 2021, 11, 381. [Google Scholar] [CrossRef] [PubMed]
- Giaretta, J.E.; Duan, H.; Oveissi, F.; Farajikhah, S.; Dehghani, F.; Naficy, S. Flexible sensors for hydrogen peroxide detection: A critical review. ACS Appl. Mater. Interfaces 2022, 14, 20491–20505. [Google Scholar] [CrossRef]
- Gupta, U.; Gupta, V.; Arun, R.K.; Chanda, N. Recent advances in enzymatic biosensors for point-of-care detection of biomolecules. Biotechnol. Bioeng. 2022, 119, 3393–3407. [Google Scholar] [CrossRef]
- Yu, Y.; Pan, M.; Peng, J.R.; Hu, D.R.; Hao, Y.; Qian, Z.Y. A review on recent advances in hydrogen peroxide electrochemical sensors for applications in cell detection. Chin. Chem. Lett. 2022, 33, 4133–4145. [Google Scholar] [CrossRef]
- Calabria, D.; Trozzi, I.; Lazzarini, E.; Pace, A.; Zangheri, M.; Iannascoli, L.; Davis, N.M.; Matadha, S.S.G.; Albuquerque, T.B.D.; Pirrotta, S.; et al. AstroBio-CubeSat: A lab-in-space for chemiluminescence-based astrobiology experiments. Biosens. Bioelectron. 2023, 226, 115110. [Google Scholar] [CrossRef]
- Neumann, B.; Wollenberger, U. Electrochemical biosensors employing natural and artificial heme peroxidases on semiconductors. Sensors 2020, 20, 3692. [Google Scholar] [CrossRef]
- Gao, Y.F.; Wang, Y.P.; Wang, Y.Z.; Magaud, P.; Liu, Y.T.; Zeng, F.; Yang, J.J.; Baldas, L.; Song, Y.J. Nanocatalysis meets microfluidics: A powerful platform for sensitive bioanalysis. Trends Analyt. Chem. 2023, 158, 116887. [Google Scholar] [CrossRef]
- Tummala, S.; Bandi, R.; Ho, Y.P. Synthesis of Cu-doped carbon dot/chitosan film composite as a catalyst for the colorimetric detection of hydrogen peroxide and glucose. Microchim. Acta 2022, 189, 284. [Google Scholar] [CrossRef]
- Huang, W.; Xu, Y.; Wang, Z.P.; Liao, K.; Zhang, Y.; Sun, Y.M. Dual nanozyme based on ultrathin 2D conductive MOF nanosheets intergraded with gold nanoparticles for electrochemical biosensing of H2O2 in cancer cells. Talanta 2022, 249, 123612. [Google Scholar] [CrossRef]
- Wang, M.J.; Zhang, J.B.; Zhou, X.B.; Sun, H.L.; Su, X.G. Fluorescence sensing strategy for xanthine assay based on gold nanoclusters and nanozyme. Sens. Actuators B Chem. 2022, 358, 131488. [Google Scholar] [CrossRef]
- Smutok, O.; Kavetskyy, T.; Prokopiv, T.; Serkiz, R.; Sausa, O.; Novak, I.; Svajdlenkova, H.; Mat’ko, I.; Gonchar, M.; Katz, E. Biosensor based on peroxidase-mimetic nanozyme and lactate oxidase for accurate L-lactate analysis in beverages. Biosensors 2022, 12, 1042. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, A.; Haag, R.; Schedler, U. Hydrogels and their role in biosensing applications. Adv. Healthc. Mater. 2021, 10, 2100062. [Google Scholar] [CrossRef] [PubMed]
- Vollmecke, K.; Afroz, R.; Bierbach, S.; Brenker, L.J.; Frucht, S.; Glass, A.; Giebelhaus, R.; Hoppe, A.; Kanemaru, K.; Lazarek, M.; et al. Hydrogel-based biosensors. Gels 2022, 8, 768. [Google Scholar] [CrossRef] [PubMed]
- Calabria, D.; Guardigli, M.; Severi, P.; Trozzi, I.; Pace, A.; Cinti, S.; Zangheri, M.; Mirasoli, M. A Smartphone-based chemosensor to evaluate antioxidants in agri-food matrices by in situ AuNP formation. Sensors 2021, 21, 5432. [Google Scholar] [CrossRef]
- Shafique, H.; de Vries, J.; Strauss, J.; Jahromi, A.K.; Moakhar, R.S.; Mahshid, S. Advances in the translation of electrochemical hydrogel-based sensors. Adv. Healthc. Mater. 2023, 12, 2201501. [Google Scholar] [CrossRef] [PubMed]
- Gacanin, J.; Synatschke, C.V.; Weil, T. Biomedical applications of DNA-based hydrogels. Adv. Funct. Mater. 2020, 30, 1906253. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.Y.; Duan, J.; Liu, J.W.; Yi, H.; Zhang, Z.Z.; Song, H.W.; Li, Y.C.; Zhang, K.X. Stimuli-responsive self-degradable DNA hydrogels: Design, synthesis, and applications. Adv. Healthc. Mater. 2023; In press. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lu, N.; Peng, H.; Li, J.; Yan, R.; Shi, X.; Ma, P.; Lv, M.; Wang, L.; Tang, Z.; et al. Multi-triggered and enzyme-mimicking graphene oxide/polyvinyl alcohol/G-quartet supramolecular hydrogels. Nanoscale 2020, 12, 5186–5195. [Google Scholar] [CrossRef]
- Bhattacharyya, T.; Kumar, Y.P.; Dash, J. Supramolecular hydrogel inspired from DNA structures mimics peroxidase activity. ACS Biomater. Sci. Eng. 2017, 3, 2358–2365. [Google Scholar] [CrossRef]
- Lena, S.; Masiero, S.; Pieraccini, S.; Spada, G.P. Guanosine hydrogen-bonded scaffolds: A new way to control the bottom-up realisation of well-defined nanoarchitectures. Chem. Eur. J. 2009, 15, 7792–7806. [Google Scholar] [CrossRef]
- Pieraccini, S.; Campitiello, M.; Carducci, F.; Davis, J.T.; Mariani, P.; Masiero, S. Playing supramolecular dominoes with light: Building and breaking a photoreversible G-quadruplex made from guanosine, boric acid and an azobenzene. Org. Biomol. Chem. 2019, 17, 2759–2769. [Google Scholar] [CrossRef]
- Peters, G.M.; Skala, L.P.; Plank, T.N.; Hyman, B.J.; Manjunatha Reddy, G.N.; Marsh, A.; Brown, S.P.; Davis, J.T. A G4·K+ hydrogel stabilized by an anion. J. Am. Chem. Soc. 2014, 136, 12596–12599. [Google Scholar] [CrossRef] [Green Version]
- Peters, G.M.; Skala, L.P.; Plank, T.N.; Oh, H.; Manjunatha Reddy, G.N.; Marsh, A.; Brown, S.P.; Raghavan, S.R.; Davis, J.T. G4-quartet·M+ borate hydrogels. J. Am. Chem. Soc. 2015, 137, 5819–5827. [Google Scholar] [CrossRef] [Green Version]
- Plank, T.N.; Davis, J.T. A G4·K+ hydrogel that self-destructs. Chem. Commun. 2016, 52, 5037–5040. [Google Scholar] [CrossRef]
- Zhong, R.; Tang, Q.; Wang, S.; Zhang, H.; Zhang, F.; Xiao, M.; Man, T.; Qu, X.; Li, L.; Zhang, W.; et al. Self-assembly of enzyme-like nanofibrous G-molecular hydrogel for printed flexible electrochemical sensors. Adv. Mater. 2018, 30, 1706887. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.H.; Vyas, C.; Grieve, B.; Bartolo, P. Recent advances in enzymatic and non-enzymatic electrochemical glucose sensing. Sensors 2021, 21, 4672. [Google Scholar] [CrossRef] [PubMed]
- Adeel, M.; Rahman, M.; Caligiuri, I.; Canzonieri, V.; Rizzolio, F.; Daniele, S. Recent advances of electrochemical and optical en-zyme-free glucose sensors operating at physiological conditions. Biosens. Bioelectron. 2020, 165, 112331. [Google Scholar] [CrossRef] [PubMed]
- Hwang, D.-W.; Lee, S.; Seo, M.; Chung, T.D. Recent advances in electrochemical non-enzymatic glucose sensors—A review. Anal. Chim. Acta 2018, 1033, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhao, Y.; Liu, Y. Current development in wearable glucose meters. Chin. Chem. Lett. 2021, 32, 3705–3717. [Google Scholar] [CrossRef]
- Yoneda, J.S.; de Araujo, D.R.; Sella, F.; Liguori, G.R.; Liguori, T.T.A.; Moreira, L.F.P.; Spinozzi, F.; Mariani, P.; Itri, R. Self-assembled guanosine-hydrogels for drug-delivery application: Structural and mechanical characterization, methylene blue loading and controlled release. Mater. Sci. Eng. C 2021, 121, 111834. [Google Scholar] [CrossRef]
- Yu, Y.; Nakamura, D.; DeBoyace, K.; Neisius, A.W.; McGown, L.B. Tunable thermoassociation of binary guanosine gels. J. Phys. Chem. B 2008, 112, 1130–1134. [Google Scholar] [CrossRef]
- Zangheri, M.; Di Nardo, F.; Calabria, D.; Marchegiani, E.; Anfossi, L.; Guardigli, M.; Mirasoli, M.; Baggiani, C.; Roda, A. Smartphone biosensor for point-of-need chemiluminescence detection of ochratoxin A in wine and coffee. Anal. Chim. Acta 2021, 1163, 338515. [Google Scholar] [CrossRef]
- Pierre, A.C. The sol-gel encapsulation of enzymes. Biocatal. Biotransformation 2004, 22, 145–170. [Google Scholar] [CrossRef]
- Ye, J.; Zhu, L.; Yan, M.; Xiao, T.; Fan, L.; Xue, Y.; Huang, J.; Yang, X. An intensive and glow-type chemiluminescence of luminol-embedded, guanosine-derived hydrogel. Talanta 2021, 230, 122351. [Google Scholar] [CrossRef]
- Basiaga, M.; Paszenda, Z.; Walke, W.; Karasiński, P.; Marciniak, J. Electrochemical impedance spectroscopy and corrosion resistance of SiO2 coated cpTi and Ti-6Al-7Nb alloy. In Information Technologies in Biomedicine; Piętka, E., Kawa, J., Wieclawek, W., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; Volume 4, pp. 411–420. [Google Scholar] [CrossRef]
- Calabria, D.; Lazzarini, E.; Pace, A.; Trozzi, I.; Zangheri, M.; Cinti, S.; Difonzo, M.; Valenti, G.; Guardigli, M.; Paolucci, F.; et al. Smartphone-based 3D-printed electrochemiluminescence enzyme biosensor for reagentless glucose quantification in real matrices. Biosens. Bioelectron. 2023, 227, 115146. [Google Scholar] [CrossRef] [PubMed]
- Baldassarri, E.J.; Ortore, M.G.; Spinozzi, F.; Round, A.; Ferrero, C.; Mariani, P. K+ vs. Na+ effects on the self-assembly of guanosine 5’-monophosphate: A solution SAXS structural study. Nanomaterials 2020, 10, 629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weibel, M.K.; Bright, H.J. The glucose oxidase mechanism: Interpretation of the pH dependence. J. Biol. Chem. 1971, 246, 2734–2744. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, T.; Biswas, A.; Gavel, P.K.; Das, A.K. Engineered dynamic boronate ester-mediated self-healable biocompatible G-quadruplex hydrogels for sustained release of vitamins. Langmuir 2020, 36, 1574–1584. [Google Scholar] [CrossRef]
- Forman, H.J.; Bernardo, A.; Davies, K.J.A. What is the concentration of hydrogen peroxide in blood and plasma? Arch. Biochem. Biophys. 2016, 603, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Ma, J.N.; Neng, J.; Yang, B.L.; Wang, Y.; Xing, F.G. A novel and label-free chemiluminescence detection of zearalenone based on a truncated aptamer conjugated with a G-quadruplex DNAzyme. Biosensors 2023, 13, 118. [Google Scholar] [CrossRef]
- Cheng, M.; Cheng, Y.; Hao, J.; Jia, G.; Zhou, J.; Mergny, J.-L.; Li, C. Loop permutation affects the topology and stability of G-quadruplexes. Nucleic Acids Res. 2018, 46, 9264–9275. [Google Scholar] [CrossRef] [Green Version]
- Calabria, D.; Mirasoli, M.; Guardigli, M.; Simoni, P.; Zangheri, M.; Severi, P.; Caliceti, C.; Roda, A. Paper-based smartphone chemosensor for reflectometric on-site total polyphenols quantification in olive oil. Sens. Actuators B Chem. 2020, 305, 127522. [Google Scholar] [CrossRef]
- McDonald, J.H. Handbook of Biological Statistics, 3rd ed.; Sparky House Publishing: Baltimore, MA, USA, 2014; pp. 222–230. [Google Scholar]
- Snedecor, G.W.; Cochran, W.G. Statistical Methods, 8th ed.; Iowa State University Press: Ames, IA, USA, 1989; pp. 374–397. [Google Scholar]
- Danaei, G.; Finucane, M.M.; Lu, Y.; Singh, G.M.; Cowan, M.J.; Paciorek, C.J.; Lin, J.K.; Farzadfar, F.; Khang, Y.H.; Stevens, G.A.; et al. National, regional, and global trends in fasting plasma glucose and diabetes prevalence since 1980: Systematic analysis of health examination surveys and epidemiological studies with 370 country-years and 2·7 million participants. Lancet 2011, 378, 31–40. [Google Scholar] [CrossRef]








| Glucose Concentration (mmol L−1) | Smartphone-Based CL Biosensor 2 | Colorimetric Reference Method 2 | ||||
|---|---|---|---|---|---|---|
| Found (mmol L−1) | Recovery (%) | RSD (%) | Found (mmol L−1) | Recovery (%) | RSD (%) | |
| 0.5 | 0.55 ± 0.05 | 110 | 9.1 | 0.45 ± 0.10 | 90 | 22.2 |
| 3.0 | 3.25 ± 0.15 | 108 | 4.6 | 2.90 ± 0.15 | 97 | 5.2 |
| 5.0 | 4.80 ± 0.30 | 96 | 6.3 | 5.25 ± 0.40 | 105 | 7.6 |
| 6.5 | 6.00 ± 0.50 | 92 | 8.3 | 6.50 ± 0.50 | 100 | 7.7 |
| 8.0 | 8.20 ± 1.00 | 103 | 12.2 | 7.40 ± 0.65 | 93 | 8.8 |
| 10.0 | 10.7 ± 0.80 | 107 | 7.4 | 9.45 ± 0.55 | 95 | 5.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calabria, D.; Pace, A.; Lazzarini, E.; Trozzi, I.; Zangheri, M.; Guardigli, M.; Pieraccini, S.; Masiero, S.; Mirasoli, M. Smartphone-Based Chemiluminescence Glucose Biosensor Employing a Peroxidase-Mimicking, Guanosine-Based Self-Assembled Hydrogel. Biosensors 2023, 13, 650. https://doi.org/10.3390/bios13060650
Calabria D, Pace A, Lazzarini E, Trozzi I, Zangheri M, Guardigli M, Pieraccini S, Masiero S, Mirasoli M. Smartphone-Based Chemiluminescence Glucose Biosensor Employing a Peroxidase-Mimicking, Guanosine-Based Self-Assembled Hydrogel. Biosensors. 2023; 13(6):650. https://doi.org/10.3390/bios13060650
Chicago/Turabian StyleCalabria, Donato, Andrea Pace, Elisa Lazzarini, Ilaria Trozzi, Martina Zangheri, Massimo Guardigli, Silvia Pieraccini, Stefano Masiero, and Mara Mirasoli. 2023. "Smartphone-Based Chemiluminescence Glucose Biosensor Employing a Peroxidase-Mimicking, Guanosine-Based Self-Assembled Hydrogel" Biosensors 13, no. 6: 650. https://doi.org/10.3390/bios13060650
APA StyleCalabria, D., Pace, A., Lazzarini, E., Trozzi, I., Zangheri, M., Guardigli, M., Pieraccini, S., Masiero, S., & Mirasoli, M. (2023). Smartphone-Based Chemiluminescence Glucose Biosensor Employing a Peroxidase-Mimicking, Guanosine-Based Self-Assembled Hydrogel. Biosensors, 13(6), 650. https://doi.org/10.3390/bios13060650