Development of an Innovative Colorimetric DNA Biosensor Based on Sugar Measurement
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Apparatus
2.3. Microplate-Based Phenol–Sulfuric Acid Method for Sugar Source Analysis
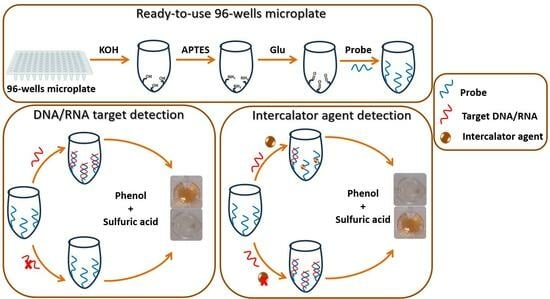
2.4. Construction of the Phenol–Sulfuric Acid-Method-Based Developed Biosensor for DNA or RNA Target Detection
2.4.1. Activation and Functional Modification of 96-Well Microplate Surfaces
2.4.2. Immobilizing Probe on the Amine-Functionalized 96-Well Microplate
2.4.3. Target RNA/DNA Hybridization
2.4.4. Target DNA/RNA Detection via Phenol–Sulfuric Acid Method
2.5. Construction of a Biosensor for Intercalating Agent Detection Based on the Phenol–Sulfuric Acid Method
3. Results and Discussions
3.1. Integrated Approach for Sugar Source Analysis and DNA Detection Using Microplate-Based Phenol–Sulfuric Acid Method
3.1.1. Sugar Reaction via the Developed Phenol–Sulfuric Acid Method
3.1.2. Application of the Phenol–Sulfuric Acid Sugar Reaction Method for DNA of Fish Sperm Analysis
3.1.3. Application of the Phenol–Sulfuric Acid Sugar Reaction Method for Each Component of DNA

3.2. DNA-Based Biosensor for Specific Targets (DNA and RNA) Detection Using the Phenol–Sulfuric Acid Method
3.3. Optimizing Phenol and Sulfuric Acid Concentration Parameters for the Developed Biosensor Based on the Phenol–Sulfuric Acid reaction
3.4. Application of the Developed DNA-Based Biosensor for DNA or RNA Target Detection
3.4.1. Analytical Performance
3.4.2. Selectivity
3.5. Application of the DNA-Based Biosensor Developed for the Detection of Intercalating Agents
Application for Curcumin Intercalating Agent

4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Masuko, T.; Minami, A.; Iwasaki, N.; Majima, T.; Nishimura, S.-I.; Lee, Y.C. Carbohydrate analysis by a phenol–sulfuric acid method in microplate format. Anal. Biochem. 2005, 1, 69–72. [Google Scholar] [CrossRef]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. A colorimetric method for the determination of sugars. Nature 1951, 1, 167. [Google Scholar] [CrossRef]
- DuBois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 1, 350–356. [Google Scholar] [CrossRef]
- Laurentin, A.; Edwards, C.A. A microtiter modification of the anthrone-sulfuric acid colorimetric assay for glucose-based carbohydrates. Anal. Biochem. Biochem. 2003, 1, 143–145. [Google Scholar] [CrossRef]
- Irwin, M.; Leaver, A.G. Use of the orcinol-sulphuric acid reaction in the positive identification of certain monosaccharides from a salivary mucoid. Nature 1956, 1, 1126. [Google Scholar] [CrossRef]
- Monsigny, M.; Petit, C.; Roche, A.-C. Colorimetric determination of neutral sugars by a resorcinol sulfuric acid micromethod. Anal. Biochem. 1988, 1, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Zhang, C. Electrogenerated chemiluminescence biosensing. Anal. Chem. 2019, 1, 524–534. [Google Scholar] [CrossRef] [PubMed]
- El Aamri, M.; Yammouri, G.; Mohammadi, H.; Amine, A.; Korri-Youssoufi, H. Electrochemical biosensors for detection of microRNA as a cancer biomarker: Pros and cons. Biosensors 2020, 1, 186. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.-F.; Zhao, K.-R.; Liu, Z.-J.; Wang, L.; Ye, S.-Y.; Liang, G.-X. Cas12a-based electrochemiluminescence biosensor for target amplification-free DNA detection. Biosens. Bioelectron. 2021, 1, 112954. [Google Scholar] [CrossRef]
- Li, M.; Liu, M.; Ma, C.; Shi, C. Rapid DNA detection and one-step RNA detection catalyzed by Bst DNA polymerase and narrow-thermal-cycling. Analyst 2020, 1, 5118–5122. [Google Scholar] [CrossRef]
- Reveguk, Z.V.; Pomogaev, V.A.; Kapitonova, M.A.; Buglak, A.A.; Kononov, A.I. Structure and formation of luminescent centers in light-up Ag cluster-based DNA probes. J. Phys. Chem. C 2021, 1, 3542–3552. [Google Scholar] [CrossRef]
- Aamri, M.E.; Mohammadi, H.; Amine, A. Novel label-free colorimetric and electrochemical detection for MiRNA-21 based on the complexation of molybdate with phosphate. Microchem. J. 2022, 1, 107851. [Google Scholar] [CrossRef]
- Aamri, M.E.; Mohammadi, H.; Amine, A. Paper-Based Colorimetric Detection of miRNA-21 Using Pre-Activated Nylon Membrane and Peroxidase-Mimetic Activity of Cysteamine-Capped Gold Nanoparticles. Biosensors 2023, 13, 74. [Google Scholar] [CrossRef] [PubMed]
- Moustakim, H.; Mohammadi, H.; Amine, A. Electrochemical DNA Biosensor Based on Immobilization of a Non-Modified ssDNA Using Phosphoramidate-Bonding Strategy and Pencil Graphite Electrode Modified with AuNPs/CB and Self-Assembled Cysteamine Monolayer. Sensors 2022, 1, 9420. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, H.; Amine, A. Spectrophotometric and electrochemical determination of MicroRNA-155 using sandwich hybridization magnetic beads. Anal. Lett. 2018, 1, 411–423. [Google Scholar] [CrossRef]
- Yammouri, G.; Mohammadi, H.; Amine, A. A highly sensitive electrochemical biosensor based on carbon black and gold nanoparticles modified pencil graphite electrode for microRNA-21 detection. Chem. Afr. 2019, 1, 291–300. [Google Scholar] [CrossRef]
- Chahri, I.; Karrat, A.; Mohammadi, H.; Amine, A. Development of a New Route for the Immobilization of Unmodified Single-Stranded DNA on Chitosan Beads and Detection of Released Guanine after Hydrolysis. Molecules 2023, 1, 2088. [Google Scholar] [CrossRef]
- Alexandre, I.; Hamels, S.; Dufour, S.; Collet, J.; Zammatteo, N.; De Longueville, F.; Gala, J.-L.; Remacle, J. Colorimetric silver detection of DNA microarrays. Anal. Biochem. 2001, 1, 1–8. [Google Scholar] [CrossRef]
- Jazayeri, M.H.; Aghaie, T.; Avan, A.; Vatankhah, A.; Ghaffari, M.R.S. Colorimetric detection based on gold nano particles (GNPs): An easy, fast, inexpensive, low-cost and short time method in detection of analytes (protein, DNA, and ion). Sens. Bio-Sens. Res. 2018, 20, 1–8. [Google Scholar] [CrossRef]
- Xu, X.; Daniel, W.L.; Wei, W.; Mirkin, C.A. Colorimetric Cu2+ detection using DNA-modified gold-nanoparticle aggregates as probes and click chemistry. Small 2010, 1, 623–626. [Google Scholar] [CrossRef]
- Trinh, K.H.; Kadam, U.S.; Rampogu, S.; Cho, Y.; Yang, K.-A.; Kang, C.H.; Lee, K.-W.; Lee, K.O.; Chung, W.S.; Hong, J.C. Development of novel fluorescence-based and label-free noncanonical G4-quadruplex-like DNA biosensor for facile, specific, and ultrasensitive detection of fipronil. J. Hazard. Mater. 2022, 1, 127939. [Google Scholar] [CrossRef]
- Lucarelli, F.; Palchetti, I.; Marrazza, G.; Mascini, M. Electrochemical DNA biosensor as a screening tool for the detection of toxicants in water and wastewater samples. Talanta 2002, 1, 949–957. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, K.; Kumar, P.S. Target-receptive structural switching of ssDNA as selective and sensitive biosensor for subsequent detection of toxic Pb2+ and organophosphorus pesticide. Chemosphere 2022, 1, 132163. [Google Scholar] [CrossRef]
- Shen, L.; Wang, P.; Ke, Y. DNA nanotechnology-based biosensors and therapeutics. Adv. Healthc. Mater. 2021, 1, 2002205. [Google Scholar] [CrossRef] [PubMed]
- Untiveros, K.L.; da Silva, E.G.; de Abreu, F.C.; da Silva-Júnior, E.F.; de Araújo-Junior, J.X.; de Aquino, T.M.; Armas, S.M.; de Moura, R.O.; Mendonça-Junior, F.J.; Serafim, V.L.; et al. An electrochemical biosensor based on Hairpin-DNA modified gold electrode for detection of DNA damage by a hybrid cancer drug intercalation. Biosens. Bioelectron. 2019, 1, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.; Alwan, M.; Alamir, H.T.A.; Alkaaby, H.H.C.; Farhan, S.S.; Awadh, S.A.; Altimari, U.S.; Al-Baghdady, H.F.A.; Kadhim, A.A.; Qasim, M.T.; et al. Detection of abemaciclib, an anti-breast cancer agent, using a new electrochemical DNA biosensor. Front. Chem. 2022, 1, 980162. [Google Scholar] [CrossRef]
- Nielsen, S.S. Phenol-sulfuric acid method for total carbohydrates. In Food Analysis Laboratory Manual; Springer: Berlin/Heidelberg, Germany, 2010; pp. 47–53. [Google Scholar]
- Yue, F.; Zhang, J.; Xu, J.; Niu, T.; Lü, X.; Liu, M. Effects of monosaccharide composition on quantitative analysis of total sugar content by phenol-sulfuric acid method. Front. Nutr. 2022, 1, 963318. [Google Scholar] [CrossRef]
- Wright, K.; de Silva, K.; Purdie, A.C.; Plain, K.M. Comparison of methods for miRNA isolation and quantification from ovine plasma. Sci. Rep. 2020, 10, 825. [Google Scholar] [CrossRef]






| Nucleic Acid | Sequence (5′-3′) |
|---|---|
| Target microRNA-21 | 5′-UAGCUUAUCAGACUGAUGUUGA-3′ |
| DNAprobe (complementary sequence of microRNA-21) | 5′-AAATCAACATCAGTCTGATAAGCTA-3′ |
| Target DNA (E. coli) | 5′-TATTAACTTTACTCCCTTCCTCCCCGCTGA-3′ |
| DNAprobe (complementary sequence of DNAE. coli) | 5′-TCAGCGGGGAGGAAGGGAGTAAAGTTAATA-3′ |
| MicroRNA-155 (non-complementary oligonucleotide) | 5′-UUAAUGCUAAUCGUGAUAGGGGUU-3′ |
| Target DNA (Hepatitis B virus HBV) | 5′-GTGTCTGCGGCGTTTTATCATCTTC-3′ |
| DNAprobe (complementary sequence of DNAHBV) | 5′-TGATAAAACGCCGCAGACAC-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El Aamri, M.; Khalki, Y.; Mohammadi, H.; Amine, A. Development of an Innovative Colorimetric DNA Biosensor Based on Sugar Measurement. Biosensors 2023, 13, 853. https://doi.org/10.3390/bios13090853
El Aamri M, Khalki Y, Mohammadi H, Amine A. Development of an Innovative Colorimetric DNA Biosensor Based on Sugar Measurement. Biosensors. 2023; 13(9):853. https://doi.org/10.3390/bios13090853
Chicago/Turabian StyleEl Aamri, Maliana, Yasmine Khalki, Hasna Mohammadi, and Aziz Amine. 2023. "Development of an Innovative Colorimetric DNA Biosensor Based on Sugar Measurement" Biosensors 13, no. 9: 853. https://doi.org/10.3390/bios13090853
APA StyleEl Aamri, M., Khalki, Y., Mohammadi, H., & Amine, A. (2023). Development of an Innovative Colorimetric DNA Biosensor Based on Sugar Measurement. Biosensors, 13(9), 853. https://doi.org/10.3390/bios13090853