BCAbox Algorithm Expands Capabilities of Raman Microscope for Single Organelles Assessment
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
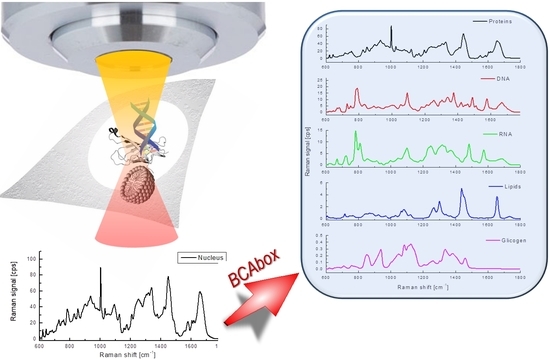
3.1. Raman Spectroscopy and BCA Toolbox Software
3.2. Results of BCA in Cellular Organelles
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Puppels, G.J.; de Mul, F.F.; Otto, C.; Greve, J.; Robert-Nicoud, M.; Arndt-Jovin, D.J.; Jovin, T.M. Studying single living cells and chromosomes by confocal Raman microspectroscopy. Nature 1990, 347, 301–303. [Google Scholar] [CrossRef] [PubMed]
- Matthaus, C.; Chernenko, T.; Newmark, J.A.; Warner, C.M.; Diem, M. Label-free detection of mitochondrial distribution in cells by nonresonant Raman microspectroscopy. Biophys. J. 2007, 93, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.J.; Zhukovsky, N.; Cass, A.E.; Nagy, J.M. Proteomics, nanotechnology and molecular diagnostics. Proteomics 2008, 8, 715–730. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Xie, Y.; Mrozek, M.F.; Ortiz, C.; Davisson, V.J.; Ben-Amotz, D. Raman detection of proteomic analytes. Anal. Chem. 2003, 75, 5703–5709. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, S.; Singh, A.K. Single cell genome sequencing. Curr. Opin. Biotechnol. 2012, 23, 437–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cacciatore, S.; Loda, M. Innovation in metabolomics to improve personalized healthcare. Ann. N. Y. Acad. Sci. 2015, 1346, 57–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.W.; Volponi, J.V.; Oliver, A.E.; Parikh, A.N.; Simmons, B.A.; Singh, S. In vivo lipidomics using single-cell Raman spectroscopy. Proc. Natl. Acad. Sci. USA 2011, 108, 3809–3814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuzmin, A.N.; Pliss, A.; Prasad, P.N. Ramanomics: New Omics Disciplines Using Micro Raman Spectrometry with Biomolecular Component Analysis for Molecular Profiling of Biological Structures. Biosensors 2017, 7, 52. [Google Scholar] [CrossRef] [PubMed]
- Pliss, A.; Kuzmin, A.N.; Kachynski, A.V.; Prasad, P.N. Nonlinear Optical Imaging and Raman Microspectrometry of the Cell Nucleus throughout the Cell Cycle. Biophys. J. 2010, 99, 3483–3491. [Google Scholar] [CrossRef] [PubMed]
- Kuzmin, A.N.; Pliss, A.; Prasad, P.N. Changes in biomolecular profile in a single nucleolus during cell fixation. Anal. Chem. 2014, 86, 10909–10916. [Google Scholar] [CrossRef] [PubMed]
- Kuzmin, A.N.; Pliss, A.; Lim, C.K.; Heo, J.; Kim, S.; Rzhevskii, A.; Gu, B.; Yong, K.T.; Wen, S.C.; Prasad, P.N. Resonance Raman Probes for Organelle-Specific Labeling in Live Cells. Sci. Rep. 2016, 6, 28483. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Heo, J.; Lim, C.K.; Pliss, A.; Kachynski, A.V.; Kuzmin, A.N.; Kim, S.; Prasad, P.N. Organelle specific imaging in live cells and immuno-labeling using resonance Raman probe. Biomaterials 2015, 53, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Pliss, A.; Kuzmin, A.N.; Kachynski, A.V.; Baev, A.; Berezney, R.; Prasad, P.N. Fluctuations and synchrony of RNA synthesis in nucleoli. Integr. Biol. 2015, 7, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Kuzmin, A.N.; Levchenko, S.M.; Pliss, A.; Qu, J.L.; Prasad, P.N. Molecular profiling of single organelles for quantitative analysis of cellular heterogeneity. Sci. Rep. 2017, 7, 6512. [Google Scholar] [CrossRef] [PubMed]
- Pliss, A.; Kuzmin, A.N.; Kachynski, A.V.; Jiang, H.B.; Hu, Z.X.; Ren, Y.; Peng, J.; Prasad, P.N. Nucleolar Molecular Signature of Pluripotent Stem Cells. Anal. Chem. 2013, 85, 3545–3552. [Google Scholar] [CrossRef] [PubMed]
- Yadav, N.; Pliss, A.; Kuzmin, A.; Rapali, P.; Sun, L.; Prasad, P.; Chandra, D. Transformations of the macromolecular landscape at mitochondria during DNA-damage-induced apoptotic cell death. Cell Death Dis. 2014, 5, e1453. [Google Scholar] [CrossRef] [PubMed]
- O’Malley, J.; Kumar, R.; Kuzmin, A.N.; Pliss, A.; Yadav, N.; Balachandar, S.; Wang, J.M.; Attwood, K.; Prasad, P.N.; Chandra, D. Lipid quantification by Raman microspectroscopy as a potential biomarker in prostate cancer. Cancer Lett. 2017, 397, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Hanlon, E.B.; Manoharan, R.; Koo, T.W.; Shafer, K.E.; Motz, J.T.; Fitzmaurice, M.; Kramer, J.R.; Itzkan, I.; Dasari, R.R.; Feld, M.S. Prospects for in vivo Raman spectroscopy. Phys. Med. Biol. 2000, 45, R1–R59. [Google Scholar] [CrossRef] [PubMed]
- Handwerger, K.E.; Cordero, J.A.; Gall, J.G. Cajal bodies, nucleoli, and speckles in the Xenopus oocyte nucleus have a low-density, sponge-like structure. Mol. Biol. Cell 2005, 16, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Pliss, A.; Peng, X.; Liu, L.X.; Kuzmin, A.; Wang, Y.; Qu, J.L.; Li, Y.; Prasad, P.N. Single Cell Assay for Molecular Diagnostics and Medicine: Monitoring Intracellular Concentrations of Macromolecules by Two-photon Fluorescence Lifetime Imaging. Theranostics 2015, 5, 919–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nolin, F.; Michel, J.; Wortham, L.; Tchelidze, P.; Balossier, G.; Banchet, V.; Bobichon, H.; Lalun, N.; Terryn, C.; Ploton, D. Changes to cellular water and element content induced by nucleolar stress: Investigation by a cryo-correlative nano-imaging approach. Cell. Mol. Life Sci. 2013, 70, 2383–2394. [Google Scholar] [CrossRef] [PubMed]
- Pliss, A.; Kuzmin, A.N.; Kachynski, A.V.; Prasad, P.N. Biophotonic probing of macromolecular transformations during apoptosis. Proc. Natl. Acad. Sci. USA 2010, 107, 12771–12776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuzmin, A.N.; Pliss, A.; Kachynski, A.V. Biomolecular component analysis of cultured cell nucleoli by Raman microspectrometry. J. Raman Spectrosc. 2013, 44, 198–204. [Google Scholar] [CrossRef]
- Levchenko, S.M.; Kuzmin, A.N.; Pliss, A.; Qu, J.; Prasad, P.N. Macromolecular Profiling of Organelles in Normal Diploid and Cancer Cells. Anal. Chem. 2017, 89, 10985–10990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czamara, K.; Majzner, K.; Pacia, M.Z.; Kochan, K.; Kaczor, A.; Baranska, M. Raman spectroscopy of lipids: A review. J. Raman Spectrosc. 2015, 46, 4–20. [Google Scholar] [CrossRef]
- Horvath, S.E.; Daum, G. Lipids of mitochondria. Prog. Lipid Res. 2013, 52, 590–614. [Google Scholar] [CrossRef] [PubMed]
- Hobro, A.J.; Rouhi, M.; Blanch, E.W.; Conn, G.L. Raman and Raman optical activity (ROA) analysis of RNA structural motifs in Domain I of the EMCV IRES. Nucleic Acids Res. 2007, 35, 1169–1177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadeghijorabchi, H.; Wilson, R.H.; Belton, P.S.; Edwardswebb, J.D.; Coxon, D.T. Quantitative-Analysis of Oils and Fats by Fourier-Transform Raman-Spectroscopy. Spectrochim. Acta A 1991, 47, 1449–1458. [Google Scholar] [CrossRef]






| Proteins | DNA | RNA | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| # of Cells | Live | F.F. | E.F. | Live | F.F. | E.F. | Live | F.F. | E.F. | |
| Nucleus | 39 | 104.6 ± 11.23 | 63.0 ± 4.9 | 67.9 ± 5.5 | 12.5 ± 4.0 | 9.3 ± 3.2 | 13.9 ± 3. | 11.6 ± 2.0 | 6.0 ± 1.9 | 5.6 ± 1.2 |
| Nucleolus | 58 | 119.1 ± 13.2 | 92.3 ± 8.0 | 114.0 ± 26.8 | 1.9 ± 2.5 | 4.1 ± 2.4 | 4.8 ± 3.2 | 34.4 ± 6.5 | 23.4 ± 4.4 | 37.2 ± 7.3 |
| M | 38 | 100.0 ± 15.5 | 76.3 ± 10.1 | 91.9 ± 12.5 | 7.4 ± 3.2 | 6.5 ± 3.2 | 10.7 ± 7.6 | 8.5 ± 2.4 | 7.2 ± 2.4 | 5.7 ± 1.0 |
| ER | 38 | 89.1 ± 11.2 | 76.5 ± 12.0 | - | 0.2 ± 0.1 | 0.3 ± 0.1 | - | 9.1 ± 3.3 | 10.0 ± 3.1 | - |
| AG | 35 | 111.5 ± 14.0 | 79.6 ± 13.1 | - | 0.2 ± 0.1 | 0.3 ± 0.2 | - | 9.3 ± 2.2 | 7.7 ± 2.5 | - |
| M.Cyt. 1 | 16 | 92.1 ± 17.5 | 54.3 ± 8.3 | 83.4 ± 23.1 | 0.7 ± 0.7 | 1.7 ± 1.6 | 0 | 12.2 ± 2.76 | 6.3 ± 1.7 | 7.9 ± 6.3 |
| M.Ch. 2 | 18 | 98.1 ± 14.4 | 51.9 ± 9.0 | 67.3 ± 22.1 | 20.8 ± 8.5 | 19.7 ± 3.9 | 30.5 ± 10.5 | 14.6 ± 2.9 | 5.0 ± 1.9 | 8.5 ± 3.9 |
| Live | F.F. | E.F. | |||||
|---|---|---|---|---|---|---|---|
| # of Cells | Concentration | 1665/1440 | Concentration | 1665/1440 | Concentration | 1665/1440 | |
| Nucleus | 39 | 4.5 ± 3.5 | 0.62 ± 0.11 | 2.0 ± 3.5 | - | 0.1 ± 0.4 | - |
| Nucleolus | 58 | 3.1 ± 3.6 | 0.65 ± 0.10 | 0.6 ± 2.1 | - | 0.1 ± 0.9 | - |
| M | 38 | 15.5 ± 12.1 | 0.57 ± 0.10 | 16.7 ± 6.5 | 0.58 ± 0.10 | 7.0 ± 10.0 | 0.63 ± 0.10 |
| ER | 38 | 23.1 ± 6.5 | 0.54 ± 0.06 | 21.2 ± 7.3 | 0.55 ± 0.07 | - | - |
| AG | 35 | 35.4 ± 10.2 | 0.43 ± 0.05 | 41.5 ± 18.2 | 0.49 ± 0.05 | - | - |
| M.Cyt. | 16 | 13.6 ± 3.5 | 0.53 ± 0.10 | 11.1 ± 9.5 | 0.51 ± 0.10 | 0 | - |
| M.Ch. | 18 | 6.2 ± 1.4 | 0.52 ± 0.10 | 4.3 ± 2.7 | 0.46 ± 0.06 | 3.0 ± 3.0 | 0.74 ± 0.06 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuzmin, A.N.; Pliss, A.; Rzhevskii, A.; Lita, A.; Larion, M. BCAbox Algorithm Expands Capabilities of Raman Microscope for Single Organelles Assessment. Biosensors 2018, 8, 106. https://doi.org/10.3390/bios8040106
Kuzmin AN, Pliss A, Rzhevskii A, Lita A, Larion M. BCAbox Algorithm Expands Capabilities of Raman Microscope for Single Organelles Assessment. Biosensors. 2018; 8(4):106. https://doi.org/10.3390/bios8040106
Chicago/Turabian StyleKuzmin, Andrey N., Artem Pliss, Alex Rzhevskii, Adrian Lita, and Mioara Larion. 2018. "BCAbox Algorithm Expands Capabilities of Raman Microscope for Single Organelles Assessment" Biosensors 8, no. 4: 106. https://doi.org/10.3390/bios8040106
APA StyleKuzmin, A. N., Pliss, A., Rzhevskii, A., Lita, A., & Larion, M. (2018). BCAbox Algorithm Expands Capabilities of Raman Microscope for Single Organelles Assessment. Biosensors, 8(4), 106. https://doi.org/10.3390/bios8040106