Regulation of Host Defense Peptide Synthesis by Polyphenols
Abstract
:1. Introduction
2. Host Defense Peptides
2.1. Classifications of HDPs
2.2. Mechanism of Antimicrobial Action of HDPs
2.3. Role of HDPs in Innate and Adaptive Immunity
2.4. Transcriptional Regulation of HDPs
3. Polyphenols
3.1. Classifications of Polyphenols
3.2. Antioxidant and Anti-Inflammatory Activities of Polyphenols
3.3. Role of Polyphenols in Cancer, Cardiovascular, and Neurological Diseases
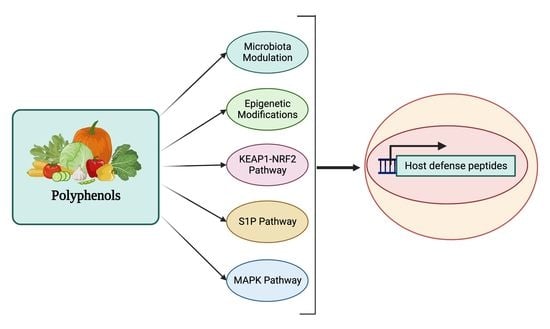
4. Regulation of Host Defense Peptides by Polyphenols
4.1. Regulation of HDPs by Flavonols
4.2. Regulation of HDPs by Flavones
4.3. Regulation of HDPs by Isoflavones
4.4. Regulation of HDPs by Flavanols
4.5. Regulation of HDPs by Anthocyanins
4.6. Regulation of HDPs by Phenolic Acids
4.7. Regulation of HDPs by Stilbenes
4.8. Regulation of HDPs by Other Polyphenols
| Class | Polyphenol | Model | HDPs Affected 1 | References |
|---|---|---|---|---|
| Flavonols | Quercetin | Human HepG2 hepatocytes | ↑ HAMP | [54] |
| Rat liver | ↑ HAMP | [54] | ||
| Zebrafish liver | ↑ defensin, LEAP2 | [55] | ||
| Silkworm larvae | ↑ defensin and cecropin A | [56] | ||
| Chicken HTC macrophages | ↑ AvBD1, AvBD2, AvBD3, AvBD4, AvBD5, AvBD6, AvBD7, AvBD9, AvBD14 | [57] | ||
| Chicken ileum | ↑ AvBD3, AvBD4, AvBD6, AvBD7, AvBD9, AvBD10, AvBD11 | [58] | ||
| Flavones | Flavone | Human SW620 colon epithelial | ↑ CAMP | [59] |
| Chrysin | Human A549 lung epithelial | ↑ CAMP | [60] | |
| Human keratinocytes stimulated with TNF-α, IL-17A, or IL-22 | ↓ S100A7, S100A8, S100A9, DEFB2, CAMP | [64] | ||
| Saponarin | Human HaCaT keratinocytes | ↑ CAMP, ↓ DEFB1 | [61] | |
| Apigenin | Human HaCaT keratinocytes | ↑ DEFB1, DEFB2, DEFB3, CAMP | [62] | |
| Mouse epidermis | ↑ Camp, Defb3 | [63] | ||
| Isoflavones | Genistein | Human LNCaP prostate cancer | ↑ DEFB1 | [65] |
| Human HaCaT keratinocytes | ↑ CAMP | [66] | ||
| Porcine endometrial epithelial | ↑ PBD2, PBD3 | [67,68] | ||
| Daidzein | Porcine endometrial epithelial | ↑ PBD2, PBD3 | [67,68] | |
| Calycosin | Porcine 3D4/31 macrophages | ↑ PBD2, PEP2C, PG1-5 | [39] | |
| Porcine IPEC-J2 intestinal epithelial | ↑ PBD2, PEP2C, PBD3 | [39] | ||
| Porcine jejunal explants | ↑ PBD3 | [39] | ||
| Flavanols | Epigallocatechin gallate | Human B11 oral epithelial | ↑ DEFB1, DEFB2 | [69] |
| Human OBA-9 gingival epithelial | ↑ DEFB1, DEFB2 | [70] | ||
| BEAS-2B bronchial epithelial | ↑ DEFB3 | [71] | ||
| Chicken PBMC | ↑ AvBD9 | [57] | ||
| Porcine IPEC-J2 intestinal | ↑ PBD1, PBD2 | [72] | ||
| Human Caco-2 colonic epithelial stimulated with Tegafur | ↑ DEFA5, DEFA6 | [73] | ||
| Antho- cyanins | Cyanidin 3-O-glycoside | Mouse intestinal mucosa | ↑ total β-defensin protein | [74] |
| Blueberry powder | Rat ileum | ↑ Defb2 | [75] | |
| Grape pomace extract | Mouse colon | ↑ REG3G | [76] | |
| Delphinidin | Human psoriatic keratinocytes | ↓ S100A7, S100A15 | [77] | |
| Phenolic Acids | Ellagic acid | Human gingival epithelial | ↑ DEFB2 | [78] |
| Caffeic acid phenethyl ester | Mouse tongue | ↑ Defb3 | [79] | |
| Galleria mellonella larvae | ↑ galiomycin, galeriomycin | [79] | ||
| Anacardic acid | Chicken PBMC | ↑ AvBD9 | [57] | |
| Stilbenes | Resveratrol | Human U937 monocytes | ↑ CAMP | [80] |
| Human HaCaT keratinocytes | ↑ CAMP | [80] | ||
| Mouse epidermis | ↑ CAMP | [81] | ||
| Mouse ileum | ↑ Defa3, Defa5, Defa20 | [82] | ||
| Human HepG2 hepatocytes | ↑ HAMP | [54] | ||
| Rat liver | ↑ HAMP | [54] | ||
| Chicken PBMC | ↑ AvBD9 | [57] | ||
| Streptococcus pneumonia-infected human A549 lung epithelial | ↑ DEFB2 | [83] | ||
| Pseudomonas aeruginosa-infected A549 lung epithelial | ↓ DEFB2 | [84,85] | ||
| Pterostilbene | Porcine 3D4/31 macrophages | ↑ PBD3, PG1-5 | [39] | |
| Pinostilbene | Porcine 3D4/31 macrophages | ↑ PBD3, PG1-5 | [39] | |
| Isorhapontigenin | Porcine IPEC-J2 intestinal epithelial | ↑ PBD3, PEP2C, PG1-5 | [39] | |
| Porcine jejunal explants | ↑ PBD3, PG1-5 | [39] | ||
| Curcuminoid | Curcumin | Human U937 monocytes | ↑ CAMP | [86] |
| Human HT29 colonic epithelial | ↑ CAMP | [86] | ||
| Grass carp liver and blood | ↑ DEFB1, HAMP, LEAP2 | [87] | ||
| Tannins | Condensed tannins | Grass carp intestine | ↓ HAMP, LEAP2A, LEAP2B, DEFB1 | [88] |
| Flavanones | Naringenin | Human HepG2 hepatocytes | ↑ HAMP | [54] |
| Chalcones | Isoliquiritigenin | Human Caco-2 colonic epithelial | ↑ DEFB3 | [90] |
| Xanthohumol | Porcine IPEC-J2 intestinal | ↑ PEP2C, PBD3, PG1-5 | [39] | |
| Porcine 3D4/31 macrophages | ↑ PEP2C, PBD3, PG1-5 | [39] | ||
| Porcine jejunal explants | ↑ PBD3, PG1-5 | [39] | ||
| Others | Gossypol | Grass carp intestine | ↓ HAMP, LEAP2B, DEFB1 | [89] |
| Deoxyshikonin | Porcine IPEC-J2 intestinal epithelial Porcine 3D4/31 macrophages Porcine jejunal explants | ↑ PEP2C, PBD3, PG1-5 ↑ PEP2C, PBD3, PG1-5 ↑ PBD3, PG1-5 | [39] [39] [39] | |
| Pinto beans | Mouse ileum | ↑ REG3B, REG3G | [92] | |
| Gold kiwifruit | Rat colon | ↑ Defb1, Defb2 | [93] |
5. Mechanism of Action for Polyphenols
5.1. HDP Regulation by the MAPK Signaling Pathway
5.2. HDP Regulation by the S1P Signaling Pathway
5.3. HDP Regulation by the KEAP1-NRF2 Pathway
5.4. HDP Regulation through Epigenetic Modifications
5.4.1. Inhibition of Histone Acetylation
5.4.2. Inhibition of DNA and Histone Methylation
5.5. HDP Regulation through Modulation of Gut Microbial Metabolites
6. Future Prospects and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Watkins, R.R.; Bonomo, R.A. Overview: The Ongoing Threat of Antimicrobial Resistance. Infect. Dis. Clin. N. Am. 2020, 34, 649–658. [Google Scholar] [CrossRef]
- Schrader, S.M.; Vaubourgeix, J.; Nathan, C. Biology of antimicrobial resistance and approaches to combat it. Sci. Transl. Med. 2020, 12, eaaz6992. [Google Scholar] [CrossRef] [PubMed]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef] [PubMed]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef] [PubMed]
- Dijksteel, G.S.; Ulrich, M.M.W.; Middelkoop, E.; Boekema, B.K.H.L. Review: Lessons Learned From Clinical Trials Using Antimicrobial Peptides (AMPs). Front. Microbiol. 2021, 12, 616979. [Google Scholar] [CrossRef]
- Rodrigues, G.; Souza Santos, L.; Franco, O.L. Antimicrobial Peptides Controlling Resistant Bacteria in Animal Production. Front. Microbiol. 2022, 13, 874153. [Google Scholar] [CrossRef]
- Bergman, P.; Raqib, R.; Rekha, R.S.; Agerberth, B.; Gudmundsson, G.H. Host Directed Therapy Against Infection by Boosting Innate Immunity. Front. Immunol. 2020, 11, 1209. [Google Scholar] [CrossRef]
- Chen, J.; Zhai, Z.; Long, H.; Yang, G.; Deng, B.; Deng, J. Inducible expression of defensins and cathelicidins by nutrients and associated regulatory mechanisms. Peptides 2020, 123, 170177. [Google Scholar] [CrossRef]
- Rodriguez-Carlos, A.; Jacobo-Delgado, Y.M.; Santos-Mena, A.O.; Rivas-Santiago, B. Modulation of cathelicidin and defensins by histone deacetylase inhibitors: A potential treatment for multi-drug resistant infectious diseases. Peptides 2021, 140, 170527. [Google Scholar] [CrossRef]
- Robinson, K.; Ma, X.; Liu, Y.; Qiao, S.; Hou, Y.; Zhang, G. Dietary modulation of endogenous host defense peptide synthesis as an alternative approach to in-feed antibiotics. Anim. Nutr. 2018, 4, 160–169. [Google Scholar] [CrossRef]
- Thakur, A.; Sharma, A.; Alajangi, H.K.; Jaiswal, P.K.; Lim, Y.B.; Singh, G.; Barnwal, R.P. In pursuit of next-generation therapeutics: Antimicrobial peptides against superbugs, their sources, mechanism of action, nanotechnology-based delivery, and clinical applications. Int. J. Biol. Macromol. 2022, 218, 135–156. [Google Scholar] [CrossRef] [PubMed]
- Alford, M.A.; Baquir, B.; Santana, F.L.; Haney, E.F.; Hancock, R.E.W. Cathelicidin Host Defense Peptides and Inflammatory Signaling: Striking a Balance. Front. Microbiol. 2020, 11, 1902. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Lu, W. Defensins: A Double-Edged Sword in Host Immunity. Front. Immunol. 2020, 11, 764. [Google Scholar] [CrossRef] [PubMed]
- Selsted, M.E.; Ouellette, A.J. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 2005, 6, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Pazgier, M.; Jung, G.; Nuccio, S.P.; Castillo, P.A.; de Jong, M.F.; Winter, M.G.; Winter, S.E.; Wehkamp, J.; Shen, B.; et al. Human alpha-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science 2012, 337, 477–481. [Google Scholar] [CrossRef] [Green Version]
- Gennaro, R.; Zanetti, M.; Benincasa, M.; Podda, E.; Miani, M. Pro-rich antimicrobial peptides from animals: Structure, biological functions and mechanism of action. Curr. Pharm. Des. 2002, 8, 763–778. [Google Scholar] [CrossRef]
- Cardoso, M.H.; Meneguetti, B.T.; Costa, B.O.; Buccini, D.F.; Oshiro, K.G.N.; Preza, S.L.E.; Carvalho, C.M.E.; Migliolo, L.; Franco, O.L. Non-Lytic Antibacterial Peptides That Translocate Through Bacterial Membranes to Act on Intracellular Targets. Int. J. Mol. Sci. 2019, 20, 4877. [Google Scholar] [CrossRef] [Green Version]
- Sass, V.; Schneider, T.; Wilmes, M.; Körner, C.; Tossi, A.; Novikova, N.; Shamova, O.; Sahl, H.-G. Human beta-Defensin 3 Inhibits Cell Wall Biosynthesis in Staphylococci. Infect. Immun. 2010, 78, 2793–2800. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.; Chen, Q.; Schmidt, A.P.; Anderson, G.M.; Wang, J.M.; Wooters, J.; Oppenheim, J.J.; Chertov, O. Ll-37, the Neutrophil Granule–And Epithelial Cell–Derived Cathelicidin, Utilizes Formyl Peptide Receptor–Like 1 (Fprl1) as a Receptor to Chemoattract Human Peripheral Blood Neutrophils, Monocytes, and T Cells. J. Exp. Med. 2000, 192, 1069–1074. [Google Scholar] [CrossRef] [Green Version]
- Rohrl, J.; Yang, D.; Oppenheim, J.J.; Hehlgans, T. Human beta-defensin 2 and 3 and their mouse orthologs induce chemotaxis through interaction with CCR2. J. Immunol. 2010, 184, 6688–6694. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.; Chertov, O.; Bykovskaia, S.N.; Chen, Q.; Buffo, M.J.; Shogan, J.; Anderson, M.; Schroder, J.M.; Wang, J.M.; Howard, O.M.; et al. Beta-defensins: Linking innate and adaptive immunity through dendritic and T cell CCR6. Science 1999, 286, 525–528. [Google Scholar] [CrossRef]
- Petrov, V.; Funderburg, N.; Weinberg, A.; Sieg, S. Human β defensin-3 induces chemokines from monocytes and macrophages: Diminished activity in cells from HIV-infected persons. Immunology 2013, 140, 413–420. [Google Scholar] [CrossRef]
- Biragyn, A.; Ruffini, P.A.; Leifer, C.A.; Klyushnenkova, E.; Shakhov, A.; Chertov, O.; Shirakawa, A.K.; Farber, J.M.; Segal, D.M.; Oppenheim, J.J.; et al. Toll-Like Receptor 4-Dependent Activation of Dendritic Cells by β-Defensin 2. Science 2002, 298, 1025–1029. [Google Scholar] [CrossRef]
- Funderburg, N.; Lederman, M.M.; Feng, Z.; Drage, M.G.; Jadlowsky, J.; Harding, C.V.; Weinberg, A.; Sieg, S.F. Human -defensin-3 activates professional antigen-presenting cells via Toll-like receptors 1 and 2. Proc. Natl. Acad. Sci. USA 2007, 104, 18631–18635. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Yu, H.; Gu, Y.; Xin, A.; Zhang, Y.; Diao, H.; Lin, D. Human beta-defensin DEFB126 is capable of inhibiting LPS-mediated inflammation. Appl. Microbiol. Biotechnol. 2013, 97, 3395–3408. [Google Scholar] [CrossRef]
- Mookherjee, N.; Brown, K.L.; Bowdish, D.M.E.; Doria, S.; Falsafi, R.; Hokamp, K.; Roche, F.M.; Mu, R.; Doho, G.H.; Pistolic, J.; et al. Modulation of the TLR-Mediated Inflammatory Response by the Endogenous Human Host Defense Peptide LL-37. J. Immunol. 2006, 176, 2455–2464. [Google Scholar] [CrossRef] [Green Version]
- Contreras, G.; Shirdel, I.; Braun, M.S.; Wink, M. Defensins: Transcriptional regulation and function beyond antimicrobial activity. Dev. Comp. Immunol. 2020, 104, 103556. [Google Scholar] [CrossRef]
- Lyu, W.; Curtis, A.R.; Sunkara, L.T.; Zhang, G. Transcriptional Regulation of Antimicrobial Host Defense Peptides. Curr. Protein Pept. Sci. 2015, 16, 672–679. [Google Scholar] [CrossRef]
- Wehkamp, K.; Schwichtenberg, L.; Schröder, J.-M.M.; Harder, J. Pseudomonas aeruginosa- and IL-1β-Mediated Induction of Human β-Defensin-2 in Keratinocytes Is Controlled by NF-κB and AP-1. J. Investig. Dermatol. 2006, 126, 121–127. [Google Scholar] [CrossRef] [Green Version]
- Gerstel, U.; Czapp, M.; Bartels, J.; Schröder, J.-M. Rhamnolipid-induced shedding of flagellin from Pseudomonas aeruginosa provokes hBD-2 and IL-8 response in human keratinocytes. Cell. Microbiol. 2009, 11, 842–853. [Google Scholar] [CrossRef]
- O’Neil, D.A.; Porter, E.M.; Elewaut, D.; Anderson, G.M.; Eckmann, L.; Ganz, T.; Kagnoff, M.F. Expression and Regulation of the Human β-Defensins hBD-1 and hBD-2 in Intestinal Epithelium. J. Immunol. 1999, 163, 6718–6724. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Sunkara, L.T.; Zeng, X.; Deng, Z.; Myers, S.M.; Zhang, G. Differential regulation of human cathelicidin LL-37 by free fatty acids and their analogs. Peptides 2013, 50, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Sunkara, L.T.; Jiang, W.; Zhang, G. Modulation of antimicrobial host defense peptide gene expression by free fatty acids. PLoS ONE 2012, 7, e49558. [Google Scholar] [CrossRef] [Green Version]
- Ren, M.; Zhang, S.; Liu, X.; Li, S.; Mao, X.; Zeng, X.; Qiao, S. Different Lipopolysaccharide Branched-Chain Amino Acids Modulate Porcine Intestinal Endogenous β-Defensin Expression through the Sirt1/ERK/90RSK Pathway. J. Agric. Food Chem. 2016, 64, 3371–3379. [Google Scholar] [CrossRef] [PubMed]
- Schlee, M.; Harder, J.; Koten, B.; Stange, E.F.; Wehkamp, J.; Fellermann, K. Probiotic lactobacilli and VSL#3 induce enterocyte beta-defensin 2. Clin. Exp. Immunol. 2008, 151, 528–535. [Google Scholar] [CrossRef]
- Lyu, W.; Deng, Z.; Zhang, G. High-Throughput Screening for Epigenetic Compounds That Induce Human beta-Defensin 1 Synthesis. Antibiotics 2023, 12, 186. [Google Scholar] [CrossRef]
- Lyu, W.; Mi, D.; Vinson, P.N.; Zhang, G. Large-scale Identification of Multiple Classes of Host Defense Peptide-Inducing Compounds for Antimicrobial Therapy. Int. J. Mol. Sci. 2022, 23, 8400. [Google Scholar] [CrossRef]
- Deng, Z.; Lyu, W.; Zhang, G. High-Throughput Identification of Epigenetic Compounds to Enhance Chicken Host Defense Peptide Gene Expression. Antibiotics 2022, 11, 933. [Google Scholar] [CrossRef]
- Wang, J.; Lyu, W.; Zhang, W.; Chen, Y.; Luo, F.; Wang, Y.; Ji, H.; Zhang, G. Discovery of natural products capable of inducing porcine host defense peptide gene expression using cell-based high throughput screening. J. Anim. Sci. Biotechnol. 2021, 12, 14. [Google Scholar] [CrossRef]
- Lyu, W.; Deng, Z.; Sunkara, L.T.; Becker, S.; Robinson, K.; Matts, R.; Zhang, G. High Throughput Screening for Natural Host Defense Peptide-Inducing Compounds as Novel Alternatives to Antibiotics. Front. Cell. Infect. Microbiol. 2018, 8, 191. [Google Scholar] [CrossRef] [Green Version]
- Nylen, F.; Miraglia, E.; Cederlund, A.; Ottosson, H.; Stromberg, R.; Gudmundsson, G.H.; Agerberth, B. Boosting innate immunity: Development and validation of a cell-based screening assay to identify LL-37 inducers. Innate Immun. 2014, 20, 364–376. [Google Scholar] [CrossRef] [Green Version]
- Rana, A.; Samtiya, M.; Dhewa, T.; Mishra, V.; Aluko, R.E. Health benefits of polyphenols: A concise review. J. Food Biochem. 2022, 46, e14264. [Google Scholar] [CrossRef]
- Durazzo, A.; Lucarini, M.; Souto, E.B.; Cicala, C.; Caiazzo, E.; Izzo, A.A.; Novellino, E.; Santini, A. Polyphenols: A concise overview on the chemistry, occurrence, and human health. Phytother. Res. 2019, 33, 2221–2243. [Google Scholar] [CrossRef] [Green Version]
- Rudrapal, M.; Khairnar, S.J.; Khan, J.; Dukhyil, A.B.; Ansari, M.A.; Alomary, M.N.; Alshabrmi, F.M.; Palai, S.; Deb, P.K.; Devi, R. Dietary Polyphenols and Their Role in Oxidative Stress-Induced Human Diseases: Insights Into Protective Effects, Antioxidant Potentials and Mechanism(s) of Action. Front. Pharmacol. 2022, 13, 806470. [Google Scholar] [CrossRef]
- Yahfoufi, N.; Alsadi, N.; Jambi, M.; Matar, C. The Immunomodulatory and Anti-Inflammatory Role of Polyphenols. Nutrients 2018, 10, 1618. [Google Scholar] [CrossRef] [Green Version]
- Chow, S.-E.; Hshu, Y.-C.; Wang, J.-S.; Chen, J.-K. Resveratrol attenuates oxLDL-stimulated NADPH oxidase activity and protects endothelial cells from oxidative functional damages. J. Appl. Physiol. 2007, 102, 1520–1527. [Google Scholar] [CrossRef] [Green Version]
- Deby-Dupont, G.; Mouithys-Mickalad, A.; Serteyn, D.; Lamy, M.; Deby, C. Resveratrol and curcumin reduce the respiratory burst of Chlamydia-primed THP-1 cells. Biochem. Biophys. Res. Commun. 2005, 333, 21–27. [Google Scholar] [CrossRef]
- Ruiz, P.A.; Braune, A.; Holzlwimmer, G.; Quintanilla-Fend, L.; Haller, D. Quercetin inhibits TNF-induced NF-kappaB transcription factor recruitment to proinflammatory gene promoters in murine intestinal epithelial cells. J. Nutr. 2007, 137, 1208–1215. [Google Scholar] [CrossRef] [Green Version]
- Sharma, E.; Attri, D.C.; Sati, P.; Dhyani, P.; Szopa, A.; Sharifi-Rad, J.; Hano, C.; Calina, D.; Cho, W.C. Recent updates on anticancer mechanisms of polyphenols. Front. Cell Dev. Biol. 2022, 10, 1005910. [Google Scholar] [CrossRef]
- Wang, S.; Du, Q.; Meng, X.; Zhang, Y. Natural polyphenols: A potential prevention and treatment strategy for metabolic syndrome. Food Funct. 2022, 13, 9734–9753. [Google Scholar] [CrossRef]
- Caruso, G.; Torrisi, S.A.; Mogavero, M.P.; Currenti, W.; Castellano, S.; Godos, J.; Ferri, R.; Galvano, F.; Leggio, G.M.; Grosso, G.; et al. Polyphenols and neuroprotection: Therapeutic implications for cognitive decline. Pharmacol. Ther. 2022, 232, 108013. [Google Scholar] [CrossRef] [PubMed]
- Chairez-Ramirez, M.H.; de la Cruz-Lopez, K.G.; Garcia-Carranca, A. Polyphenols as Antitumor Agents Targeting Key Players in Cancer-Driving Signaling Pathways. Front. Pharmacol. 2021, 12, 710304. [Google Scholar] [CrossRef] [PubMed]
- Islam, F.; Islam, M.M.; Khan Meem, A.F.; Nafady, M.H.; Islam, M.R.; Akter, A.; Mitra, S.; Alhumaydhi, F.A.; Emran, T.B.; Khusro, A.; et al. Multifaceted role of polyphenols in the treatment and management of neurodegenerative diseases. Chemosphere 2022, 307, 136020. [Google Scholar] [CrossRef] [PubMed]
- Bayele, H.K.; Balesaria, S.; Srai, S.K. Phytoestrogens modulate hepcidin expression by Nrf2: Implications for dietary control of iron absorption. Free Radic. Biol. Med. 2015, 89, 1192–1202. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhang, C.; Zhang, J.; Xie, J.; Yang, L.; Xing, Y.; Li, Z. The effects of quercetin on immunity, antioxidant indices, and disease resistance in zebrafish (Danio rerio). Fish Physiol. Biochem. 2020, 46, 759–770. [Google Scholar] [CrossRef]
- Shi, G.; Kang, Z.; Ren, F.; Zhou, Y.; Guo, P. Effects of Quercetin on the Growth and Expression of Immune-Pathway-Related Genes in Silkworm (Lepidoptera: Bombycidae). J. Insect Sci. 2020, 20, 23. [Google Scholar] [CrossRef]
- Yang, Q.; Burkardt, A.C.; Sunkara, L.T.; Xiao, K.; Zhang, G. Natural Cyclooxygenase-2 Inhibitors Synergize with Butyrate to Augment Chicken Host Defense Peptide Gene Expression. Front. Immunol. 2022, 13, 819222. [Google Scholar] [CrossRef]
- Ying, L.; Wu, H.; Zhou, S.; Lu, H.; Ding, M.; Wang, B.; Wang, S.; Mao, Y.; Xiao, F.; Li, Y. Toll-Like Receptors Signaling Pathway of Quercetin Regulating Avian Beta-Defensin in the Ileum of Broilers. Front. Cell Dev. Biol. 2022, 10, 816771. [Google Scholar] [CrossRef]
- Schauber, J.; Svanholm, C.; Termen, S.; Iffland, K.; Menzel, T.; Scheppach, W.; Melcher, R.; Agerberth, B.; Luhrs, H.; Gudmundsson, G.H. Expression of the cathelicidin LL-37 is modulated by short chain fatty acids in colonocytes: Relevance of signalling pathways. Gut 2003, 52, 735–741. [Google Scholar] [CrossRef] [Green Version]
- Xin, Y.; Chen, S.; Tang, K.; Wu, Y.; Guo, Y. Identification of Nifurtimox and Chrysin as Anti-Influenza Virus Agents by Clinical Transcriptome Signature Reversion. Int. J. Mol. Sci. 2022, 23, 2372. [Google Scholar] [CrossRef]
- Min, S.Y.; Park, C.H.; Yu, H.W.; Park, Y.J. Anti-Inflammatory and Anti-Allergic Effects of Saponarin and Its Impact on Signaling Pathways of RAW 264.7, RBL-2H3, and HaCaT Cells. Int. J. Mol. Sci. 2021, 22, 8431. [Google Scholar] [CrossRef]
- Park, C.H.; Min, S.Y.; Yu, H.W.; Kim, K.; Kim, S.; Lee, H.J.; Kim, J.H.; Park, Y.J. Effects of Apigenin on RBL-2H3, RAW264.7, and HaCaT Cells: Anti-Allergic, Anti-Inflammatory, and Skin-Protective Activities. Int. J. Mol. Sci. 2020, 21, 4620. [Google Scholar] [CrossRef]
- Hou, M.; Sun, R.; Hupe, M.; Kim, P.L.; Park, K.; Crumrine, D.; Lin, T.K.; Santiago, J.L.; Mauro, T.M.; Elias, P.M.; et al. Topical apigenin improves epidermal permeability barrier homoeostasis in normal murine skin by divergent mechanisms. Exp. Dermatol. 2013, 22, 210–215. [Google Scholar] [CrossRef] [Green Version]
- Li, H.-J.; Wu, N.-L.; Pu, C.-M.; Hsiao, C.-Y.; Chang, D.-C.; Hung, C.-F. Chrysin alleviates imiquimod-induced psoriasis-like skin inflammation and reduces the release of CCL20 and antimicrobial peptides. Sci. Rep. 2020, 10, 2932. [Google Scholar] [CrossRef] [Green Version]
- Merchant, K.; Kumi-Diaka, J.; Rathinavelu, A.; Esiobu, N.; Zoeller, R.; Hormann, V. Genistein modulation of immune-assoicated genes in LNCaP prostate cancer cell line. Open Prostate Cancer J. 2012, 5, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Park, K.; Kim, Y.I.; Shin, K.O.; Seo, H.S.; Kim, J.Y.; Mann, T.; Oda, Y.; Lee, Y.M.; Holleran, W.M.; Elias, P.M.; et al. The dietary ingredient, genistein, stimulates cathelicidin antimicrobial peptide expression through a novel S1P-dependent mechanism. J. Nutr. Biochem. 2014, 25, 734–740. [Google Scholar] [CrossRef] [Green Version]
- Srisomboon, Y.; Poonyachoti, S.; Deachapunya, C. Soy isoflavones enhance β-defensin synthesis and secretion in endometrial epithelial cells with exposure to TLR3 agonist polyinosinic-polycytidylic acid. Am. J. Reprod. Immunol. 2017, 78, e12694. [Google Scholar] [CrossRef]
- Srisomboon, Y.; Poonyachoti, S.; Deachapunya, C. Enhanced Secretion of Beta-Defensins in Endometrial Tissues and Epithelial Cells by Soy Isoflavones. J. Med. Assoc. Thai. 2016, 99 (Suppl. S8), S142–S149. [Google Scholar]
- Lombardo Bedran, T.B.; Feghali, K.; Zhao, L.; Palomari Spolidorio, D.M.; Grenier, D. Green tea extract and its major constituent, epigallocatechin-3-gallate, induce epithelial beta-defensin secretion and prevent beta-defensin degradation by Porphyromonas gingivalis. J. Periodontal Res. 2014, 49, 615–623. [Google Scholar] [CrossRef]
- Lombardo Bedran, T.B.; Morin, M.P.; Palomari Spolidorio, D.; Grenier, D. Black Tea Extract and Its Theaflavin Derivatives Inhibit the Growth of Periodontopathogens and Modulate Interleukin-8 and β-Defensin Secretion in Oral Epithelial Cells. PLoS ONE 2015, 10, e0143158. [Google Scholar] [CrossRef]
- Mou, Q.; Jiang, Y.; Zhu, L.; Zhu, Z.; Ren, T. EGCG induces β-defensin 3 against influenza A virus H1N1 by the MAPK signaling pathway. Exp. Ther. Med. 2020, 20, 3017–3024. [Google Scholar] [CrossRef] [PubMed]
- Wan, M.L.; Ling, K.H.; Wang, M.F.; El-Nezami, H. Green tea polyphenol epigallocatechin-3-gallate improves epithelial barrier function by inducing the production of antimicrobial peptide pBD-1 and pBD-2 in monolayers of porcine intestinal epithelial IPEC-J2 cells. Mol. Nutr. Food Res. 2016, 60, 1048–1058. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Kobayashi, M.; Ogura, J.; Yamaguchi, H.; Satoh, T.; Watanabe, K.; Iseki, K. Immunoprotective effect of epigallocatechin-3-gallate on oral anticancer drug-induced α-defensin reduction in Caco-2 cells. Biol. Pharm. Bull. 2014, 37, 490–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Zou, C.; Liu, Y. Amelioration of Ovalbumin-Induced Food Allergy in Mice by Targeted Rectal and Colonic Delivery of Cyanidin-3-O-Glucoside. Foods 2022, 11, 1542. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Keirsey, K.I.; Kirkland, R.; Grunewald, Z.I.; Fischer, J.G.; de La Serre, C.B. Blueberry Supplementation Influences the Gut Microbiota, Inflammation, and Insulin Resistance in High-Fat-Diet-Fed Rats. J. Nutr. 2018, 148, 209–219. [Google Scholar] [CrossRef] [Green Version]
- Van Hul, M.; Geurts, L.; Plovier, H.; Druart, C.; Everard, A.; Ståhlman, M.; Rhimi, M.; Chira, K.; Teissedre, P.L.; Delzenne, N.M.; et al. Reduced obesity, diabetes, and steatosis upon cinnamon and grape pomace are associated with changes in gut microbiota and markers of gut barrier. Am. J. Physiol. Endocrinol. Metab. 2018, 314, E334–E352. [Google Scholar] [CrossRef]
- Chamcheu, J.C.; Pal, H.C.; Siddiqui, I.A.; Adhami, V.M.; Ayehunie, S.; Boylan, B.T.; Noubissi, F.K.; Khan, N.; Syed, D.N.; Elmets, C.A.; et al. Prodifferentiation, anti-inflammatory and antiproliferative effects of delphinidin, a dietary anthocyanidin, in a full-thickness three-dimensional reconstituted human skin model of psoriasis. Skin Pharmacol. Physiol. 2015, 28, 177–188. [Google Scholar] [CrossRef] [Green Version]
- Promsong, A.; Chung, W.O.; Satthakarn, S.; Nittayananta, W. Ellagic acid modulates the expression of oral innate immune mediators: Potential role in mucosal protection. J. Oral Pathol. Med. 2015, 44, 214–221. [Google Scholar] [CrossRef]
- de Barros, P.P.; Rossoni, R.D.; Garcia, M.T.; Kaminski, V.L.; Loures, F.V.; Fuchs, B.B.; Mylonakis, E.; Junqueira, J.C. The Anti-Biofilm Efficacy of Caffeic Acid Phenethyl Ester (CAPE) In Vitro and a Murine Model of Oral Candidiasis. Front. Cell Infect. Microbiol. 2021, 11, 700305. [Google Scholar] [CrossRef]
- Guo, C.; Sinnott, B.; Niu, B.; Lowry, M.B.; Fantacone, M.L.; Gombart, A.F. Synergistic induction of human cathelicidin antimicrobial peptide gene expression by vitamin D and stilbenoids. Mol. Nutr. Food Res. 2014, 58, 528–536. [Google Scholar] [CrossRef] [Green Version]
- Park, K.; Elias, P.M.; Hupe, M.; Borkowski, A.W.; Gallo, R.L.; Shin, K.-O.; Lee, Y.-M.; Holleran, W.M.; Uchida, Y. Resveratrol Stimulates Sphingosine-1-Phosphate Signaling of Cathelicidin Production. J. Investig. Dermatol. 2013, 133, 1942–1949. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, Y.; Huang, H.; Liu, S.; Liu, F.; Tu, Q.; Yin, Y.; He, S. Resveratrol Improves Growth Performance, Intestinal Morphology, and Microbiota Composition and Metabolism in Mice. Front. Microbiol. 2021, 12, 726878. [Google Scholar] [CrossRef]
- Lin, L.; Wen, S.H.; Guo, S.Z.; Su, X.Y.; Wu, H.J.; Chong, L.; Zhang, H.L.; Zhang, W.X.; Li, C.C. Role of SIRT1 in Streptococcus pneumoniae-induced human β-defensin-2 and interleukin-8 expression in A549 cell. Mol. Cell. Biochem. 2014, 394, 199–208. [Google Scholar] [CrossRef]
- Cerqueira, A.M.; Khaper, N.; Lees, S.J.; Ulanova, M. The antioxidant resveratrol down-regulates inflammation in an in-vitro model of Pseudomonas aeruginosa infection of lung epithelial cells. Can. J. Physiol. Pharmacol. 2013, 91, 248–255. [Google Scholar] [CrossRef] [Green Version]
- Euba, B.; López-López, N.; Rodríguez-Arce, I.; Fernández-Calvet, A.; Barberán, M.; Caturla, N.; Martí, S.; Díez-Martínez, R.; Garmendia, J. Resveratrol therapeutics combines both antimicrobial and immunomodulatory properties against respiratory infection by nontypeable Haemophilus influenzae. Sci. Rep. 2017, 7, 12860. [Google Scholar] [CrossRef] [Green Version]
- Guo, C.; Rosoha, E.; Lowry, M.B.; Borregaard, N.; Gombart, A.F. Curcumin induces human cathelicidin antimicrobial peptide gene expression through a vitamin D receptor-independent pathway. J. Nutr. Biochem. 2013, 24, 754–759. [Google Scholar] [CrossRef] [Green Version]
- Ming, J.; Ye, J.; Zhang, Y.; Xu, Q.; Yang, X.; Shao, X.; Qiang, J.; Xu, P. Optimal dietary curcumin improved growth performance, and modulated innate immunity, antioxidant capacity and related genes expression of NF-κB and Nrf2 signaling pathways in grass carp (Ctenopharyngodon idella) after infection with Aeromonas hydrophila. Fish Shellfish Immunol. 2020, 97, 540–553. [Google Scholar] [CrossRef]
- Li, M.; Feng, L.; Jiang, W.D.; Wu, P.; Liu, Y.; Jiang, J.; Kuang, S.Y.; Tang, L.; Zhou, X.Q. Condensed tannins decreased the growth performance and impaired intestinal immune function in on-growing grass carp (Ctenopharyngodon idella). Br. J. Nutr. 2020, 123, 737–755. [Google Scholar] [CrossRef]
- Wang, K.-Z.; Feng, L.; Jiang, W.-D.; Wu, P.; Liu, Y.; Jiang, J.; Kuang, S.-Y.; Tang, L.; Zhang, Y.-A.; Zhou, X.-Q. Dietary gossypol reduced intestinal immunity and aggravated inflammation in on-growing grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol. 2019, 86, 814–831. [Google Scholar] [CrossRef]
- Sechet, E.; Telford, E.; Bonamy, C.; Sansonetti, P.J.; Sperandio, B. Natural molecules induce and synergize to boost expression of the human antimicrobial peptide β-defensin-3. Proc. Natl. Acad. Sci. USA 2018, 115, E9869–E9878. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.-Q.; Gan, R.-Y.; Ge, Y.-Y.; Zhang, D.; Corke, H. Polyphenols in Common Beans (Phaseolus vulgaris L.): Chemistry, Analysis, and Factors Affecting Composition. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1518–1539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ojo, B.A.; Lu, P.; Alake, S.E.; Keirns, B.; Anderson, K.; Gallucci, G.; Hart, M.D.; El-Rassi, G.D.; Ritchey, J.W.; Chowanadisai, W.; et al. Pinto beans modulate the gut microbiome, augment MHC II protein, and antimicrobial peptide gene expression in mice fed a normal or western-style diet. J. Nutr. Biochem. 2021, 88, 108543. [Google Scholar] [CrossRef] [PubMed]
- Paturi, G.; Butts, C.A.; Bentley-Hewitt, K.L.; Ansell, J. Influence of green and gold kiwifruit on indices of large bowel function in healthy rats. J. Food Sci. 2014, 79, H1611–H1620. [Google Scholar] [CrossRef] [PubMed]
- Rezatabar, S.; Karimian, A.; Rameshknia, V.; Parsian, H.; Majidinia, M.; Kopi, T.A.; Bishayee, A.; Sadeghinia, A.; Yousefi, M.; Monirialamdari, M.; et al. RAS/MAPK signaling functions in oxidative stress, DNA damage response and cancer progression. J. Cell. Physiol. 2019, 234, 14951–14965. [Google Scholar] [CrossRef] [PubMed]
- Maceyka, M.; Harikumar, K.B.; Milstien, S.; Spiegel, S. Sphingosine-1-phosphate signaling and its role in disease. Trends Cell Biol. 2012, 22, 50–60. [Google Scholar] [CrossRef] [Green Version]
- Park, K.; Elias, P.M.; Shin, K.O.; Lee, Y.M.; Hupe, M.; Borkowski, A.W.; Gallo, R.L.; Saba, J.; Holleran, W.M.; Uchida, Y. A novel role of a lipid species, sphingosine-1-phosphate, in epithelial innate immunity. Mol. Cell. Biol. 2013, 33, 752–762. [Google Scholar] [CrossRef] [Green Version]
- Deshmukh, P.; Unni, S.; Krishnappa, G.; Padmanabhan, B. The Keap1-Nrf2 pathway: Promising therapeutic target to counteract ROS-mediated damage in cancers and neurodegenerative diseases. Biophys. Rev. 2017, 9, 41–56. [Google Scholar] [CrossRef] [Green Version]
- Balogun, E.; Hoque, M.; Gong, P.; Killeen, E.; Green, C.J.; Foresti, R.; Alam, J.; Motterlini, R. Curcumin activates the haem oxygenase-1 gene via regulation of Nrf2 and the antioxidant-responsive element. Biochem. J. 2003, 371, 887–895. [Google Scholar] [CrossRef] [Green Version]
- Yao, J.; Zhang, B.; Ge, C.; Peng, S.; Fang, J. Xanthohumol, a Polyphenol Chalcone Present in Hops, Activating Nrf2 Enzymes to Confer Protection against Oxidative Damage in PC12 Cells. J. Agric. Food Chem. 2015, 63, 1521–1531. [Google Scholar] [CrossRef]
- Portela, A.; Esteller, M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010, 28, 1057–1068. [Google Scholar] [CrossRef]
- Perri, F.; Longo, F.; Giuliano, M.; Sabbatino, F.; Favia, G.; Ionna, F.; Addeo, R.; Della Vittoria Scarpati, G.; Di Lorenzo, G.; Pisconti, S. Epigenetic control of gene expression: Potential implications for cancer treatment. Crit. Rev. Oncol. Hematol. 2017, 111, 166–172. [Google Scholar] [CrossRef]
- Verdone, L.; Caserta, M.; Mauro, E.D. Role of histone acetylation in the control of gene expression. Biochem. Cell Biol. 2005, 83, 344–353. [Google Scholar] [CrossRef]
- Bouyahya, A.; El Hachlafi, N.; Aanniz, T.; Bourais, I.; Mechchate, H.; Benali, T.; Shariati, M.A.; Burkov, P.; Lorenzo, J.M.; Wilairatana, P.; et al. Natural Bioactive Compounds Targeting Histone Deacetylases in Human Cancers: Recent Updates. Molecules 2022, 27, 2568. [Google Scholar] [CrossRef]
- Majid, S.; Dar, A.A.; Shahryari, V.; Hirata, H.; Ahmad, A.; Saini, S.; Tanaka, Y.; Dahiya, A.V.; Dahiya, R. Genistein reverses hypermethylation and induces active histone modifications in tumor suppressor gene B-Cell translocation gene 3 in prostate cancer. Cancer 2010, 116, 66–76. [Google Scholar] [CrossRef] [Green Version]
- Ciesielski, O.; Biesiekierska, M.; Balcerczyk, A. Epigallocatechin-3-gallate (EGCG) Alters Histone Acetylation and Methylation and Impacts Chromatin Architecture Profile in Human Endothelial Cells. Molecules 2020, 25, 2326. [Google Scholar] [CrossRef]
- Hait, N.C.; Allegood, J.; Maceyka, M.; Strub, G.M.; Harikumar, K.B.; Singh, S.K.; Luo, C.; Marmorstein, R.; Kordula, T.; Milstien, S.; et al. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science 2009, 325, 1254–1257. [Google Scholar] [CrossRef] [Green Version]
- Mayack, B.K.; Sippl, W.; Ntie-Kang, F. Natural Products as Modulators of Sirtuins. Molecules 2020, 25, 3287. [Google Scholar] [CrossRef]
- Noh, Y.-H.; Lee, J.; Seo, S.J.; Myung, S.C. Promoter DNA methylation contributes to human β-defensin-1 deficiency in atopic dermatitis. Anim. Cells Syst. 2018, 22, 172–177. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.Q.; Arnold, R.; Fernandez-Golarz, C.; Parrish, A.B.; Almekinder, T.; He, J.; Ho, S.-m.; Svoboda, P.; Pohl, J.; Marshall, F.F.; et al. Human β-Defensin-1, a Potential Chromosome 8p Tumor Suppressor: Control of Transcription and Induction of Apoptosis in Renal Cell Carcinoma. Cancer Res. 2006, 66, 8542–8549. [Google Scholar] [CrossRef] [Green Version]
- Whitmore, M.A.; Li, H.; Lyu, W.; Khanam, S.; Zhang, G. Epigenetic Regulation of Host Defense Peptide Synthesis: Synergy Between Histone Deacetylase Inhibitors and DNA/Histone Methyltransferase Inhibitors. Front. Immunol. 2022, 13, 874706. [Google Scholar] [CrossRef]
- Saldivar-Gonzalez, F.I.; Gomez-Garcia, A.; Chavez-Ponce de Leon, D.E.; Sanchez-Cruz, N.; Ruiz-Rios, J.; Pilon-Jimenez, B.A.; Medina-Franco, J.L. Inhibitors of DNA Methyltransferases From Natural Sources: A Computational Perspective. Front. Pharmacol. 2018, 9, 1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akone, S.H.; Ntie-Kang, F.; Stuhldreier, F.; Ewonkem, M.B.; Noah, A.M.; Mouelle, S.E.M.; Muller, R. Natural Products Impacting DNA Methyltransferases and Histone Deacetylases. Front. Pharmacol. 2020, 11, 992. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Qi, Y.; Zheng, H. Dietary Polyphenol, Gut Microbiota, and Health Benefits. Antioxidants 2022, 11, 1212. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, F.; Wu, W.; Sun, M.; Bilotta, A.J.; Yao, S.; Xiao, Y.; Huang, X.; Eaves-Pyles, T.D.; Golovko, G.; et al. GPR43 mediates microbiota metabolite SCFA regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mTOR and STAT3. Mucosal Immunol. 2018, 11, 752–762. [Google Scholar] [CrossRef] [Green Version]
- Bertelli, A.; Biagi, M.; Corsini, M.; Baini, G.; Cappellucci, G.; Miraldi, E. Polyphenols: From Theory to Practice. Foods 2021, 10, 2595. [Google Scholar] [CrossRef]
- Hu, B.; Liu, X.; Zhang, C.; Zeng, X. Food macromolecule based nanodelivery systems for enhancing the bioavailability of polyphenols. J. Food Drug Anal. 2017, 25, 3–15. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Qiu, C.; Li, X.; McClements, D.J.; Jiao, A.; Wang, J.; Jin, Z. Advances in research on interactions between polyphenols and biology-based nano-delivery systems and their applications in improving the bioavailability of polyphenols. Trends. Food Sci. Technol. 2021, 116, 492–500. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tobin, I.; Zhang, G. Regulation of Host Defense Peptide Synthesis by Polyphenols. Antibiotics 2023, 12, 660. https://doi.org/10.3390/antibiotics12040660
Tobin I, Zhang G. Regulation of Host Defense Peptide Synthesis by Polyphenols. Antibiotics. 2023; 12(4):660. https://doi.org/10.3390/antibiotics12040660
Chicago/Turabian StyleTobin, Isabel, and Guolong Zhang. 2023. "Regulation of Host Defense Peptide Synthesis by Polyphenols" Antibiotics 12, no. 4: 660. https://doi.org/10.3390/antibiotics12040660
APA StyleTobin, I., & Zhang, G. (2023). Regulation of Host Defense Peptide Synthesis by Polyphenols. Antibiotics, 12(4), 660. https://doi.org/10.3390/antibiotics12040660