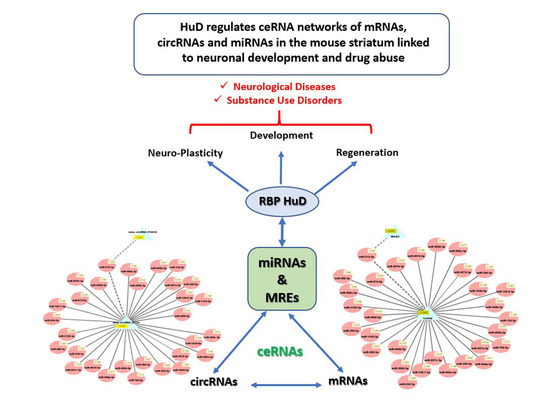
HuD Regulates mRNA-circRNA-miRNA Networks in the Mouse Striatum Linked to Neuronal Development and Drug Addiction
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Studies
2.2. RNA Extraction
2.3. mRNA Arrays and qPCR Validation
2.4. circRNA Arrays
2.5. miRNA Sequencing
2.6. Ingenuity Pathway Analysis
2.7. circRNA-miRNA-mRNA Interaction Analysis
3. Results
3.1. Alterations in mRNA Levels in the Striatum of HuD KO Mice
3.2. Altered Biological Pathways Associated with Downregulated mRNAs
3.3. Dysregulation of circRNA Levels in HuD KO Striatum
3.4. Alterations in the Levels of miRNAs in HuD KO Striatum
3.5. ceRNA Networks of miRNAs–mRNAs–circRNAs in HuD KO Striatum
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Donnelly, C.J.; Park, M.; Spillane, M.; Yoo, S.; Pacheco, A.; Gomes, C.; Vuppalanchi, D.; Mcdonald, M.; Kim, H.K.; Merianda, T.T.; et al. Axonally synthesized β-actin and GAP-43 proteins support distinct modes of axonal growth. J. Neurosci. 2013, 33, 3311–3322. [Google Scholar] [CrossRef] [Green Version]
- Holt, C.E.; Martin, K.C.; Schuman, E.M. Local translation in neurons: Visualization and function. Nat. Struct. Mol. Biol. 2019, 26, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Dalla Costa, I.; Buchanan, C.N.; Zdradzinski, M.D.; Sahoo, P.K.; Smith, T.P.; Thames, E.; Kar, A.N.; Twiss, J.L. The functional organization of axonal mRNA transport and translation. Nat. Rev. Neurosci. 2021, 22, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Campos, A.R.; And, D.G.; White, K. Mutant alleles at the locus elav in drosophila melanogaster lead to nervous system defects. A developmental-genetic analysis. J. Neurogenet. 1985, 2, 197–218. [Google Scholar] [CrossRef]
- Pascale, A.; Amadio, M.; Quattrone, A. Defining a neuron: Neuronal ELAV proteins. Cell. Mol. Life Sci. 2007, 65, 128–140. [Google Scholar] [CrossRef]
- DeBoer, E.M.; Azevedo, R.; Vega, T.A.; Brodkin, J.; Akamatsu, W.; Okano, H.; Wagner, G.C.; Rasin, M.-R. Prenatal deletion of the RNA-binding protein HuD disrupts postnatal cortical circuit maturation and behavior. J. Neurosci. 2014, 34, 3674–3686. [Google Scholar] [CrossRef] [PubMed]
- Mobarak, C.D.; Anderson, K.D.; Morin, M.; Beckel-Mitchener, A.; Rogers, S.L.; Furneaux, H.; King, P.; Perrone-Bizzozero, N.I. The RNA-binding protein HuD is required for GAP-43 mRNA stability, GAP-43 gene expression, and PKC-dependent neurite outgrowth in PC12 cells. Mol. Biol. Cell 2000, 11, 3191–3203. [Google Scholar] [CrossRef] [Green Version]
- Akten, B.; Kye, M.J.; Hao, L.T.; Wertz, M.H.; Singh, S.; Nie, D.; Huang, J.; Merianda, T.T.; Twiss, J.L.; Beattie, C.E.; et al. Interaction of survival of motor neuron (SMN) and HuD proteins with mRNA cpg15 rescues motor neuron axonal deficits. Proc. Natl. Acad. Sci. USA 2011, 108, 10337–10342. [Google Scholar] [CrossRef] [Green Version]
- Brennan, C.M.; Steitz, J.A. HuR and mRNA stability. Cell. Mol. Life Sci. CMLS 2001, 58, 266–277. [Google Scholar] [CrossRef]
- Bolognani, F.; Perrone-Bizzozero, N.I. RNA–protein interactions and control of mRNA stability in neurons. J. Neurosci. Res. 2008, 86, 481–489. [Google Scholar] [CrossRef]
- Sahoo, P.K.; Smith, D.S.; Perrone-Bizzozero, N.; Twiss, J.L. Axonal mRNA transport and translation at a glance. J. Cell Sci. 2018, 131, jcs196808. [Google Scholar] [CrossRef] [Green Version]
- Srikantan, S.; Tominaga, K.; Gorospe, M. Functional Interplay between RNA-Binding Protein HuR and microRNAs. Curr. Protein Pept. Sci. 2012, 13, 372–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolognani, F.; Merhege, M.A.; Twiss, J.; Perrone-Bizzozero, N.I. Dendritic localization of the RNA-binding protein HuD in hippocampal neurons: Association with polysomes and upregulation during contextual learning. Neurosci. Lett. 2004, 371, 152–157. [Google Scholar] [CrossRef]
- Bolognani, F.; Qiu, S.; Tanner, D.C.; Paik, J.; Perrone-Bizzozero, N.I.; Weeber, E.J. Associative and spatial learning and memory deficits in transgenic mice overexpressing the RNA-binding protein HuD. Neurobiol. Learn. Mem. 2007, 87, 635–643. [Google Scholar] [CrossRef] [PubMed]
- Pascale, A.; Gusev, P.A.; Amadio, M.; Dottorini, T.; Govoni, S.; Alkon, D.L.; Quattrone, A. Increase of the RNA-binding protein HuD and posttranscriptional up-regulation of the GAP-43 gene during spatial memory. Proc. Natl. Acad. Sci. USA 2004, 101, 1217–1222. [Google Scholar] [CrossRef] [Green Version]
- Bolognani, F.; Tanner, D.C.; Nixon, S.; Okano, H.H.J.; Okano, H.H.J.; Perrone-Bizzozero, N.I. Coordinated expression of HuD and GAP-43 in hippocampal dentate granule cells during developmental and adult plasticity. Neurochem. Res. 2007, 32, 2142–2151. [Google Scholar] [CrossRef] [PubMed]
- Tiruchinapalli, D.M.; Caron, M.G.; Keene, J.D. Activity-dependent expression of ELAV/Hu RBPs and neuronal mRNAs in seizure and cocaine brain. J. Neurochem. 2008, 107, 1529–1543. [Google Scholar] [CrossRef]
- Dell’Orco, M.; Sardone, V.; Gardiner, A.S.; Pansarasa, O.; Bordoni, M.; Perrone-Bizzozero, N.I.; Cereda, C. HuD regulates SOD1 expression during oxidative stress in differentiated neuroblastoma cells and sporadic ALS motor cortex. Neurobiol. Dis. 2021, 148, 105211. [Google Scholar] [CrossRef]
- Amadio, M.; Pascale, A.; Wang, J.; Ho, L.; Quattrone, A.; Gandy, S.; Haroutunian, V.; Racchi, M.; Pasinetti, G.M. nELAV proteins alteration in Alzheimer’s disease brain: A novel putative target for amyloid-beta reverberating on AbetaPP processing. J. Alzheimers. Dis. 2009, 16, 409–419. [Google Scholar] [CrossRef]
- Perrone-Bizzozero, N.; Bird, C.W. Role of HuD in nervous system function and pathology. Front. Biosci. (Schol. Ed). 2013, 5, 554–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeStefano, A.L.; Latourelle, J.; Lew, M.F.; Suchowersky, O.; Klein, C.; Golbe, L.I.; Mark, M.H.; Growdon, J.H.; Wooten, G.F.; Watts, R.; et al. Replication of association between ELAVL4 and Parkinson disease: The Gene PD study. Hum. Genet. 2008, 124, 95–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noureddine, M.A.; Qin, X.J.; Oliveira, S.A.; Skelly, T.J.; van der Walt, J.; Hauser, M.A.; Pericak-Vance, M.A.; Vance, J.M.; Li, Y.J. Association between the neuron-specific RNA-binding protein ELAVL4 and Parkinson disease. Hum. Genet. 2005, 117, 27–33. [Google Scholar] [CrossRef]
- Oliver, R.J.; Brigman, J.L.; Bolognani, F.; Allan, A.M.; Neisewander, J.L.; Perrone-Bizzozero, N.I. Neuronal RNA-binding protein HuD regulates addiction-related gene expression and behavior. Genes, Brain Behav. 2018, 17, 1–10. [Google Scholar] [CrossRef]
- Oliver, R.J.; Kenton, J.A.; Stevens, W.; Perrone-Bizzozero, N.I.; Brigman, J.L. Overexpression of neuronal RNA-binding protein HuD increases reward induced reinstatement of an instrumental response. Neurosci. Lett. 2018, 683, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, A.J.A.J.; Hafez, A.K.A.K.; Amoah, S.K.S.K.; Rodriguez, B.A.B.A.; Dell’Orco, M.; Lozano, E.; Hartley, B.J.B.J.; Alural, B.; Lalonde, J.; Chander, P.; et al. A psychiatric disease-related circular RNA controls synaptic gene expression and cognition. Mol. Psychiatry 2020, 25, 2712–2727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dell’Orco, M.; Oliver, R.J.; Perrone-Bizzozero, N. HuD Binds to and Regulates Circular RNAs Derived From Neuronal Development- and Synaptic Plasticity-Associated Genes. Front. Genet. 2020, 11, 796. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, A.; Twiss, J.; Perrone-Bizzozero, N. Competing Interactions of RNA-Binding Proteins, MicroRNAs, and Their Targets Control Neuronal Development and Function. Biomolecules 2015, 5, 2903–2918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Phelps, M.; Coss, C.; Wang, H.; Cook, M.; Iorns, E.; Gunn, W.; Tan, F.; Lomax, J.; Perfito, N.; Errington, T. Registered report: Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Elife 2016, 5, e12470. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.; Zhong, Z.; Lv, M.; Shu, J.; Tian, Q.; Chen, J. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget 2016, 7, 47186–47200. [Google Scholar] [CrossRef]
- Yager, L.M.; Garcia, A.F.; Wunsch, A.M.; Ferguson, S.M. The ins and outs of the striatum: Role in drug addiction. Neuroscience 2015, 301, 529–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akamatsu, W.; Fujihara, H.; Mitsuhashi, T.; Yano, M.; Shibata, S.; Hayakawa, Y.; Okano, H.J.; Sakakibara, S.-I.; Takano, H.; Takano, T.; et al. The RNA-binding protein HuD regulates neuronal cell identity and maturation. Proc. Natl. Acad. Sci. USA 2005, 102, 4625–4630. [Google Scholar] [CrossRef] [Green Version]
- PrimerBank. Available online: https://pga.mgh.harvard.edu/primerbank/ (accessed on 19 August 2021).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- MD, R.; DJ, M.; GK, S. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Ingenuity Pahtway Analysis (IPA). Available online: https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis (accessed on 18 August 2021).
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4, e05005. [Google Scholar] [CrossRef]
- TargetScan Tool at GitHub. Available online: https://github.com/vagarwal87/TargetScanTools (accessed on 13 May 2021).
- Friedman, R.C.; Farh, K.K.H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, W.; Ji, P.; Zhao, F. CircAtlas: An integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biol. 2020, 21, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Bolognani, F.; Contente-Cuomo, T.; Perrone-Bizzozero, N.I. Novel recognition motifs and biological functions of the RNA-binding protein HuD revealed by genome-wide identification of its targets. Nucleic Acids Res. 2010, 38, 117–130. [Google Scholar] [CrossRef] [Green Version]
- Merienne, N.; Cile Meunier, C.; Schneider, A.; Neri, C.; Merienne, K.; Dé Glon Correspondence, N. Cell-Type-Specific Gene Expression Profiling in Adult Mouse Brain Reveals Normal and Disease-State Signatures. Cell Rep. 2019, 26. [Google Scholar] [CrossRef] [Green Version]
- Li, C.-Y.; Mao, X.X.; Wei, L.; Nestler, E.; Uhl, G.; Goldman, D.; Oroszi, G.; Ducci, F.; Nestler, E.; Williams, K.; et al. Genes and (Common) Pathways Underlying Drug Addiction. PLoS Comput. Biol. 2008, 4, e2. [Google Scholar] [CrossRef] [Green Version]
- Vannan, A.; Powell, G.L.; Dell’Orco, M.; Wilson, M.A.; Perrone-Bizzozero, N.I.; Neisewander, J.L.; DellOrco, M.; Wilson, M.A.; Perrone-Bizzozero, N.I.; Neisewander, J.L. microRNA regulation related to the protective effects of environmental enrichment against cocaine-seeking behavior. Drug Alcohol Depend. 2021, 108585. [Google Scholar] [CrossRef]
- Hollander, J.A.; Im, H.-I.; Amelio, A.L.; Kocerha, J.; Bali, P.; Lu, Q.; Willoughby, D.; Wahlestedt, C.; Conkright, M.D.; Kenny, P.J. Striatal microRNA controls cocaine intake through CREB signalling. Nature 2010, 466, 197–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choudhury, N.R.; Michlewski, G. Terminal loop-mediated control of microRNA biogenesis. Biochem. Soc. Trans. 2012, 40, 789–793. [Google Scholar] [CrossRef] [PubMed]
- Michlewski, G.; Cáceres, J.F. Post-transcriptional control of miRNA biogenesis. RNA 2019, 25, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlezon, W.A.; Duman, R.S.; Nestler, E.J. The many faces of CREB. Trends Neurosci. 2005, 28, 436–445. [Google Scholar] [CrossRef]
- Powell, G.L.; Vannan, A.; Bastle, R.M.; Wilson, M.A.; Dell’Orco, M.; Perrone-Bizzozero, N.I.; Neisewander, J.L. Environmental enrichment during forced abstinence from cocaine self-administration opposes gene network expression changes associated with the incubation effect. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef]
- Tran-Nguyen, L.T.L.; Fuchs, R.A.; Coffey, G.P.; Baker, D.A.; O’Dell, L.E.; Neisewander, J.L. Time-dependent changes in cocaine-seeking behavior and extracellular dopamine levels in the amygdala during cocaine withdrawal. Neuropsychopharmacology 1998, 19, 48–59. [Google Scholar] [CrossRef] [Green Version]
- Im, H.I.; Hollander, J.A.; Bali, P.; Kenny, P.J. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat. Neurosci. 2010, 13, 1120–1127. [Google Scholar] [CrossRef] [Green Version]
- Sen, R.; Ghosal, S.; Das, S.; Balti, S.; Chakrabarti, J. Competing endogenous RNA: The key to posttranscriptional regulation. Sci. World J. 2014, 2014. [Google Scholar] [CrossRef]
- Cai, Y.; Wan, J. Competing Endogenous RNA Regulations in Neurodegenerative Disorders: Current Challenges and Emerging Insights. Front. Mol. Neurosci. 2018, 11. [Google Scholar] [CrossRef] [Green Version]
- Kartha, R.V.; Subramanian, S. Competing endogenous RNAs (ceRNAs): New entrants to the intricacies of gene regulation. Front. Genet. 2014, 5, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heinla, I.; Leidmaa, E.; Kongi, K.; Pennert, A.; Innos, J.; Nurk, K.; Tekko, T.; Singh, K.; Vanaveski, T.; Reimets, R.; et al. Gene expression patterns and environmental enrichment-induced effects in the hippocampi of mice suggest importance of Lsamp in plasticity. Front. Neurosci. 2015, 9, 205. [Google Scholar] [CrossRef] [Green Version]
- Innos, J.; Philips, M.A.; Raud, S.; Lilleväli, K.; Kõks, S.; Vasar, E. Deletion of the Lsamp gene lowers sensitivity to stressful environmental manipulations in mice. Behav. Brain Res. 2012, 228, 74–81. [Google Scholar] [CrossRef]
- Drewes, G.; Ebneth, A.; Preuss, U.; Mandelkow, E.M.; Mandelkow, E. MARK, a novel family of protein kinases that phosphorylate microtubule- associated proteins and trigger microtubule disruption. Cell 1997, 89, 297–308. [Google Scholar] [CrossRef]
- Lund, H.; Gustafsson, E.; Svensson, A.; Nilsson, M.; Berg, M.; Sunnemark, D.; von Euler, G. MARK4 and MARK3 associate with early tau phosphorylation in Alzheimer’s disease granulovacuolar degeneration bodies. Acta Neuropathol. Commun. 2014, 2, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Kabbaj, M.; Evans, S.; Watson, S.J.; Akil, H. The search for the neurobiological basis of vulnerability to drug abuse: Using microarrays to investigate the role of stress and individual differences. Neuropharmacology 2004, 47, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, M.W.; Kim, M.S.; Adachi, M.; Morris, M.J.; Qi, X.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N.; Kavalali, E.T.; Monteggia, L.M. In vivo analysis of mef2 transcription factors in synapse regulation and neuronal survival. PLoS One 2012, 7, 34863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, L.; She, H.; Li, W.; Zeng, J.; Zhu, J.; Jones, D.P.; Mao, Z.; Gao, G.; Yang, Q. Oxidation of survival factor MEF2D in neuronal death and Parkinson’s disease. Antioxidants Redox Signal. 2014, 20, 2936–2948. [Google Scholar] [CrossRef] [Green Version]
- Jansen, L.A.; Mirzaa, G.M.; Ishak, G.E.; O’Roak, B.J.; Hiatt, J.B.; Roden, W.H.; Gunter, S.A.; Christian, S.L.; Collins, S.; Adams, C.; et al. PI3K/AKT pathway mutations cause a spectrum of brain malformations from megalencephaly to focal cortical dysplasia. Brain 2015, 138, 1613–1628. [Google Scholar] [CrossRef] [Green Version]
- Burden-Gulley, S.M.; Brady-Kalnay, S.M. PTPμ regulates N-cadherin-dependent neurite outgrowth. J. Cell Biol. 1999, 144, 1323–1336. [Google Scholar] [CrossRef] [Green Version]
- Burden-Gulley, S.M.; Ensslen, S.E.; Brady-Kalnay, S.M. Protein Tyrosine Phosphatase-μ Differentially Regulates Neurite Outgrowth of Nasal and Temporal Neurons in the Retina. J. Neurosci. 2002, 22, 3615–3627. [Google Scholar] [CrossRef]






| Downregulated mRNAs that are upregulated in HuD OE mice and HuD targets 1 (118) | Aebp2, Ahnak, Akap8, Akap9, Anxa2, Ap3b2, Aqp9, Arf6, Arhgef10, Arhgef10l, Asph, Auts2, Axin1, Bach1, Baz1b, Bcl9, Bdnf, Bicc1, Bicd1, Brdt, Btrc, Cab39l, Cables1, Camk2a, Cfdp1, Cnn3, Col25a1, Copa, Creb1, Csnk1g1, Cyhr1, D10Wsu102e, Ddx54, Denr, Dst, Ebna1bp2, Eif2s2, Eif4g1, Eif4g3, Elavl4, Elovl5, Elovl7, Eps15, Etfdh, Ewsr1, Fndc3a, Fus, Gak, Galnt7, Gba2, Gli3, Gpm6a, Gria2, Gsk3b, Hdac9, Hells, Hmgn1, Hook2, Igf1, Impad1, Iqgap1, Iqsec2, Irf2, Kcnn2, Kif5b, Lmnb1, Lrrc19, Luc7l2, Mapk8ip3, Mcm3, Mgrn1, Mitf, Mpdz, Mpp6, Mta3, Myo1d, Nasp, Nfia, Ophn1, Paip1, Pik3r1, Plcl1, Ppp1r2, Ppwd1, Prrx1, Rab8b, Rapgef3, Rapgef5, Rbm8a, Rbms3, Rbx1, Rdh12, Rere, Rhoc, Rps6ka3, Rrm1, Scn1a, Sdha, Skil, Slc25a30, Slc8a1, Ss18, Steap1, Tardbp, Tcf4, Tfdp2, Tmpo, Tnfaip2, Tom1l2, Top1, Tpm1, Yap1, Ybx1, Zbtb7a, Zkscan1, and Zmat2 |
| Downregulated mRNAs that are also ARGs 2 (67) | Abat, Aldoc, Apod, Apoe, Atxn1, Bcl2l1, Bdnf, Btg3, Camk2a, Capzb, Cldn5, Cnp, Col1a2, Creb1, Cxcl12, Eno1, Enpp2, Erbb3, Fabp7, Fn1, Gadd45g, Gars, Gclc, Gfap, Gpm6b, Gria2, Homer2, Hspa1a, Hspa1b, Igfbp2, Jun, Kcnma1, Lamp1, Maob, Mapt, Mbp, Mobp, Mog, Mpdz, Nefl, Nfia, Nptxr, Nr4a3, Nsmaf, Ntrk2, Osbpl1a, Pam, Parp4, Pdia3, Pglyrp1, Ppp1r2, Ppp2r5c, Pura, Rad23b, Rpl23a, Scg2, Schip1, Sfpq, Slc3a2, Smpd2, Sod2, Tmed10, Tnfrsf9, Tra2a, Tubb2b, Ube2m, and Vdac1 |
| Downregulated mRNAs that are both HuD targets and ARGs 3 (32) | Abat, Aldco, Apoe, Atxn1, Bdnf, Camk2a, Col1a2, Creb1, Gria2, Cxcl12, Enpp2, Fn1, Gars, Gclc, Gfap, Gria2, Homer2, Kcnma1, Lamp1, Mpdz, Nfia, Pam, Pdia3, Ppp1r2, Pura, Rpl23a, Slc3a2, Sod2, Tra2a, Tubb2b, Ube2m, and Vdac1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dell’Orco, M.; Elyaderani, A.; Vannan, A.; Sekar, S.; Powell, G.; Liang, W.S.; Neisewander, J.L.; Perrone-Bizzozero, N.I. HuD Regulates mRNA-circRNA-miRNA Networks in the Mouse Striatum Linked to Neuronal Development and Drug Addiction. Biology 2021, 10, 939. https://doi.org/10.3390/biology10090939
Dell’Orco M, Elyaderani A, Vannan A, Sekar S, Powell G, Liang WS, Neisewander JL, Perrone-Bizzozero NI. HuD Regulates mRNA-circRNA-miRNA Networks in the Mouse Striatum Linked to Neuronal Development and Drug Addiction. Biology. 2021; 10(9):939. https://doi.org/10.3390/biology10090939
Chicago/Turabian StyleDell’Orco, Michela, Amir Elyaderani, Annika Vannan, Shobana Sekar, Gregory Powell, Winnie S. Liang, Janet L. Neisewander, and Nora I. Perrone-Bizzozero. 2021. "HuD Regulates mRNA-circRNA-miRNA Networks in the Mouse Striatum Linked to Neuronal Development and Drug Addiction" Biology 10, no. 9: 939. https://doi.org/10.3390/biology10090939
APA StyleDell’Orco, M., Elyaderani, A., Vannan, A., Sekar, S., Powell, G., Liang, W. S., Neisewander, J. L., & Perrone-Bizzozero, N. I. (2021). HuD Regulates mRNA-circRNA-miRNA Networks in the Mouse Striatum Linked to Neuronal Development and Drug Addiction. Biology, 10(9), 939. https://doi.org/10.3390/biology10090939