Relevance of miR-223 as Potential Diagnostic and Prognostic Markers in Cancer †
Abstract
:Simple Summary
Abstract
1. MicroRNAs (miRNAs)
2. General microRNA-223 Biology
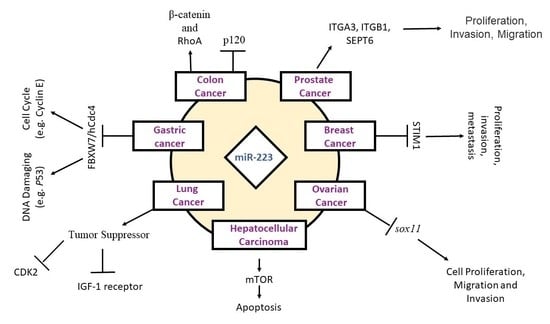
3. miR-223 and Cancer
3.1. Gastric Cancer
3.2. Hepatocellular Carcinoma
3.3. Breast Cancer
3.4. Lung Cancer
3.5. Ovarian Cancer
3.6. Osteosarcoma
3.7. Other Cancers
4. miR-223 and Key Motifs of Malignancy
4.1. Proliferation and Cell Cycle Progression Are Affected by miR-223
- 1.
- Effect of miR-223 on FOXO1 and cell proliferation
- 2.
- Regulation of IGF-1R by miR-223 to modulate cell proliferation
- 3.
- Role of miR-223 on E2F1 and cell proliferation
- 4.
- Effect of miR-223 in cytoplasmic activation/proliferation-associated protein-1 (Caprin-1)-induced cell proliferation
4.2. Metastasis and Invasion of Cancer
5. miR-223 as a Circulating Noninvasive Biomarker
6. miR-223 Application as a Diagnostic and Therapeutic Biomarker for Cancers
6.1. Hepatocellular Carcinoma and Liver Damage
6.2. Breast Cancer
6.3. Esophageal Cancer
6.4. Lung Cancer
7. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AGO | Argonaute |
| ARTN | Artemin |
| Caprin-1 | cytoplasmic activation/proliferation-associated protein-1 |
| DGCR8 | DiGeorge syndrome critical region gene 8 |
| FOXO | Forkhead box protein O |
| HCC | Hepatocellular carcinoma |
| HGSOC | high-grade serous ovarian cancer |
| IGF-1R | Insulin-like growth factor 1 receptor |
| IARC | International agency for research on cancer |
| miRNAs | microRNAs |
| mRNA | messenger RNA |
| miR-223 | microRNA-223 |
| MDSCs | Myeloid-derived suppressor cells |
| pre-miRNAs | miRNA precursors |
| RISC | RNA-induced silencing complex |
| SOX | sex determining region Y-box 11 |
| STM1 | Stathmin1 |
| UTR | Untranslated region |
References
- Hunter, M.P.; Ismail, N.; Zhang, X.; Aguda, B.D.; Lee, E.J.; Yu, L.; Xiao, T.; Schafer, J.; Lee, M.-L.T.; Schmittgen, T.D.; et al. Detection of microRNA Expression in Human Peripheral Blood Microvesicles. PLoS ONE 2008, 3, e3694. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Zhang, Y.; Zhang, H.; Liu, X.; Gong, T.; Li, M.; Sun, L.; Ji, G.; Shi, Y.; Han, Z.; et al. miRNA-223 Promotes Gastric Cancer Invasion and Metastasis by Targeting Tumor Suppressor EPB41L3. Mol. Cancer Res. 2011, 9, 824–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, L.; Chen, F. A challenge for miRNA: Multiple isomiRs in miRNAomics. Gene 2014, 544, 1–7. [Google Scholar] [CrossRef]
- Dexheimer, P.J.; Cochella, L. MicroRNAs: From Mechanism to Organism. Front. Cell Dev. Biol. 2020, 8, 409. [Google Scholar] [CrossRef]
- Haneklaus, M.; Gerlic, M.; O’Neill, L.A.J.; Masters, S.L. miR-223: Infection, inflammation and cancer. J. Intern. Med. 2013, 274, 215–226. [Google Scholar] [CrossRef]
- Ge, Q.; Brichard, S.; Yi, X.; Li, Q. microRNAs as a New Mechanism Regulating Adipose Tissue Inflammation in Obesity and as a Novel Therapeutic Strategy in the Metabolic Syndrome. J. Immunol. Res. 2014, 2014, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finnegan, E.F.; Pasquinelli, A.E. MicroRNA biogenesis: Regulating the regulators. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 51–68. [Google Scholar] [CrossRef] [Green Version]
- Aziz, F.; Yulin, L.; Yan, Q. miR-1290 stimulates proliferation of gastric cancer by targeting SP1 with overexpression of fucosyl-transferase IV and α1, 3-fucosylated glycans. biorXiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Q.; Cui, X.; Zhang, D.; Yang, Y.; Yan, X.; Liu, M.; Niang, B.; Aziz, F.; Liu, S.; Yan, Q.; et al. miR-200b inhibits proliferation and metastasis of breast cancer by targeting fucosyltransferase IV and α1,3-fucosylated glycans. Oncogenesis 2017, 6, e358. [Google Scholar] [CrossRef] [Green Version]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef]
- Finch, M.L.; Marquardt, J.U.; Yeoh, G.C.; Callus, B.A. Regulation of microRNAs and their role in liver development, regeneration and disease. Int. J. Biochem. Cell Biol. 2014, 54, 288–303. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-Z.; Li, L.; Lodish, H.F.; Bartel, D.P. MicroRNAs Modulate Hematopoietic Lineage Differentiation. Science 2004, 303, 83–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Connell, R.M.; Zhao, J.L.; Rao, D. MicroRNA function in myeloid biology. Blood 2011, 118, 2960–2969. [Google Scholar] [CrossRef] [Green Version]
- Fukao, T.; Fukuda-Yuzawa, Y.; Kiga, K.; Sharif, J.; Hino, K.; Enomoto, Y.; Kawamura, A.; Nakamura, K.; Takeuchi, T.; Tanabe, M. Matters Arising an Evolutionarily Conserved Mechanism for MicroRNA-223 Expression Revealed by MicroRNA Gene Profiling. Cell 2007, 129, 617–631. [Google Scholar] [CrossRef] [Green Version]
- Gilicze, A.B.; Wiener, Z.; Tóth, S.; Buzás, E.; Pállinger, É.; Falcone, F.; Falus, A. Myeloid-Derived microRNAs, miR-223, miR27a, and miR-652, Are Dominant Players in Myeloid Regulation. BioMed Res. Int. 2014, 2014, 870267. [Google Scholar] [CrossRef] [PubMed]
- Jeffries, J.; Zhou, W.; Hsu, A.; Deng, Q. miRNA-223 at the crossroads of inflammation and cancer. Cancer Lett. 2019, 451, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Wu, Q.; Wang, Z.; Che, Y.; Zheng, S.; Chen, Y.; Zhong, X.; Shi, F. miR-223: An Immune Regulator in Infectious Disorders. Front. Immunol. 2021, 12, 781815. [Google Scholar] [CrossRef]
- Wong, Q.W.; Lung, R.W.; Law, P.T.; Lai, P.B.-S.; Chan, K.Y.Y.; To, K.; Wong, N. MicroRNA-223 Is Commonly Repressed in Hepatocellular Carcinoma and Potentiates Expression of Stathmin1. Gastroenterology 2008, 135, 257–269. [Google Scholar] [CrossRef]
- Yendamuri, S.; Calin, G.A. The role of microRNA in human leukemia: A review. Leukemia 2009, 23, 1257–1263. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Luo, X.; Li, H.; Yue, X.; Deng, L.; Cui, Y.; Lu, Y. MicroRNA-223 functions as an oncogene in human colorectal cancer cells. Oncol. Rep. 2014, 32, 115–120. [Google Scholar] [CrossRef]
- Laios, A.; O’Toole, S.; Flavin, R.; Martin, C.; Kelly, L.; Ring, M.; Finn, S.P.; Barrett, C.; Loda, M.; Gleeson, N.; et al. Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mol. Cancer. 2008, 7, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nian, W.; Ao, X.; Wu, Y.; Huang, Y.; Shao, J.; Wang, Y.; Chen, Z.; Chen, F.; Wang, D. miR-223 functions as a potent tumor suppressor of the Lewis lung carcinoma cell line by targeting insulin-like growth factor-1 receptor and cyclin-dependent kinase 2. Oncol. Lett. 2013, 6, 359–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machado-Neto, J.A.; Teresinha, S.; Saad, O.; Traina, F. Stathmin 1 in normal and malignant hematopoiesis. BMB Rep. 2014, 47, 660–665. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Cao, T.; Cui, Y.; Zhang, F.; Shi, Y.; Xia, J.; Wang, Z.P. miR-223 Regulates Cell Proliferation and Invasion via Targeting PDS5B in Pancreatic Cancer Cells. Mol. Ther.—Nucleic Acids. 2019, 14, 583–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aziz, F. The emerging role of miR-223 as novel potential diagnostic and therapeutic target for inflammatory disorders. Cell. Immunol. 2016, 303, 1–6. [Google Scholar] [CrossRef]
- Stamatopoulos, B.; Meuleman, N.; Haibe-Kains, B.; Saussoy, P.; Van Den Neste, E.; Michaux, L.; Heimann, P.; Martiat, P.; Bron, D.; Lagneaux, L. microRNA-29c and microRNA-223 down-regulation has in vivo significance in chronic lymphocytic leukemia and improves disease risk stratification. Blood 2009, 113, 5237–5245. [Google Scholar] [CrossRef]
- Tito, C.; De Falco, E.; Rosa, P.; Iaiza, A.; Fazi, F.; Petrozza, V.; Calogero, A. Circulating microRNAs from the Molecular Mechanisms to Clinical Biomarkers: A Focus on the Clear Cell Renal Cell Carcinoma. Genes 2021, 12, 1154. [Google Scholar] [CrossRef]
- Aziz, F.; Qiu, Y. The role of anti-LeY antibody in the downregulation of MAPKs/COX-2 pathway in gastric cancer. Curr. Drug Targets. 2014, 15, 469–476. [Google Scholar] [CrossRef]
- Aziz, F.; Chakarobaty, A.; Liu, K.; Zhang, T.; Li, X.; Du, R.; Monts, J.; Xu, G.; Li, Y.; Bai, R.; et al. Gastric tumorigenesis induced by combining Helicobacter pylori infection and chronic alcohol through IL-10 inhibition. Carcinogenesis 2021, 114. [Google Scholar] [CrossRef]
- Aziz, F.; Khan, I.; Shukla, S.; Dey, D.K.; Yan, Q.; Chakraborty, A.; Yoshitomi, H.; Hwang, S.-K.; Sonwal, S.; Lee, H.; et al. Partners in crime: The Lewis Y antigen and fucosyltransferase IV in Helicobacter pylori-induced gastric cancer. Pharmacol. Ther. 2021, 107994. [Google Scholar] [CrossRef]
- Aziz, F.; Xin, M.; Gao, Y.; Chakroborty, A.; Khan, I.; Monts, J.; Monson, K.; Bode, A.M.; Dong, Z. Induction and Prevention of Gastric Cancer with Combined Helicobacter pylori and Capsaicin Administration and DFMO Treatment, Respectively. Cancers 2020, 12, 816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aziz, F.; Sherwani, S.K.; Akhtar, S.S.; Kazmi, S.U. Development of an in-house enzyme-linked immunosorbent assay based on surface whole cell antigen for diagnosis of Helicobacter pylori infection in patients with gastroduodenal ulcer disease. World J. Microbiol. Biotechnol. 2014, 30, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Aziz, F.; Yang, X.; Wen, Q.; Yan, Q. A method for establishing human primary gastric epithelial cell culture from fresh surgical gastric tissues. Mol. Med. Rep. 2015, 12, 2939–2944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aziz, F.; Taj, Y.; Kazmi, S.U. Development of an In-House Enzyme-Linked Immunosorbent Assay Based on Helicobacter pylori Sonicate Whole Cell Antigen for Diagnosis of Gastroduodenal Ulcer Disease in Karachi, Pakistan. Int. J. Microbiol. Adv. Immunol. 2013, 24–31. [Google Scholar] [CrossRef]
- Parkin, D.M.; Bray, F.; Ferlay, J.; Pisani, P. Global Cancer Statistics, 2002. CA Cancer J. Clin. 2005, 55, 74–108. [Google Scholar] [CrossRef]
- Qi, P.; Cheng, S.; Wang, H.; Li, N.; Chen, Y.; Gao, C. Serum MicroRNAs as Biomarkers for Hepatocellular Carcinoma in Chinese Patients with Chronic Hepatitis B Virus Infection. PLoS ONE 2011, 6, 6–13. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.; Shen, J.; Liu, Y.; Lin, G.; Dong, H.; Li, K. Effects of Doctor-patient Communication on Quality of Life among Breast Cancer Patients in Southern China. Asian Pac. J. Cancer Prev. 2014, 15, 5639–5644. [Google Scholar] [CrossRef] [Green Version]
- Morse, E.P.; Maegga, B.; Joseph, G.; Miesfeldt, S. Breast Cancer Knowledge, Beliefs, and Screening Practices among Women Seeking Care at District Hospitals in Dar es Salaam, Tanzania. Breast Cancer. 2014, 8, 73–79. [Google Scholar] [CrossRef] [Green Version]
- Lahart, I.M.; Metsios, G.S.; Nevill, A.M.; Carmichael, A.R. Physical Activity Levels in Women Attending Breast Screening, Receiving Chemotherapy and Post-Breast Cancer Treatment; A Cross-Sectional Study. Int. J. Environ. Res. Public Health. 2014, 11, 5487–5496. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Lin, Y.; Liu, S.; Aziz, F.; Yan, Q. Fucosyltransferase IV (FUT4) as an effective biomarker for the diagnosis of breast cancer. Biomed. Pharmacother. 2015, 70, 299–304. [Google Scholar] [CrossRef]
- Gong, B.; Hu, H.; Chen, J.; Cao, S.; Yu, J.; Xue, J.; Chen, F.; Cai, Y.; He, H.; Zhang, L. Caprin-1 is a novel microRNA-223 target for regulating the proliferation and invasion of human breast cancer cells. Biomed. Pharmacother. 2013, 67, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Rezola, A.; Pey, J.; Rubio, A.; Planes, F.J. In-Silico Prediction of Key Metabolic Differences between Two Non-Small Cell Lung Cancer Subtypes. PLoS ONE 2014, 9, e103998. [Google Scholar] [CrossRef] [PubMed]
- Martini, S.D.L.; Müller, C.B.; Meurer, R.T.; Cruz, M.; Mariano, R.; E Silva, M.B.; Klamt, F.; Andrade, C.F. The potential role of extracellular regulatory kinase in the survival of patients with early stage adenocarcinoma. J. Thorac. Dis. 2014, 6, 930–936. [Google Scholar] [CrossRef]
- Shi, J.; Jiang, X.; Yu, Z.; He, G.; Ning, H.; Wu, Z.; Cai, Y.; Yu, H.; Chen, A. ZNRF3 contributes to the growth of lung carcinoma via inhibiting Wnt/β-catenin pathway and is regulated by miR-93. Tumor Biol. 2015, 37, 3051–3057. [Google Scholar] [CrossRef]
- Chen, Z.; Wu, Y.; Meng, Q.; Xia, Z. Elevated microRNA-25 inhibits cell apoptosis in lung cancer by targeting RGS3. In Vitro Cell. Dev. Biol. Anim. 2016, 52, 62–67. [Google Scholar] [CrossRef]
- Dong, W.; Yao, C.; Teng, X.; Chai, J.; Yang, X.; Li, B. MiR-140-3p suppressed cell growth and invasion by downregulating the expression of ATP8A1 in non-small cell lung cancer. Tumor Biol. 2015, 37, 2973–2985. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2012. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [Green Version]
- Fang, G.; Liu, J.; Wang, Q.; Huang, X.; Yang, R.; Pang, Y.; Yang, M. MicroRNA-223-3p Regulates Ovarian Cancer Cell Proliferation and Invasion by Targeting SOX11 Expression. Int. J. Mol. Sci. 2017, 18, 1208. [Google Scholar] [CrossRef] [Green Version]
- Jemal, A.; Center, M.M.; DeSantis, C.; Ward, E.M. Global Patterns of Cancer Incidence and Mortality Rates and Trends. Cancer Epidemiol Biomarkers Prev. 2010, 19, 1893–1908. [Google Scholar] [CrossRef] [Green Version]
- Ji, Q.; Xu, X.; Song, Q.; Xu, Y.; Tai, Y.; Goodman, S.B.; Bi, W.; Xu, M.; Jiao, S.; Maloney, W.J.; et al. miR-223-3p Inhibits Human Osteosarcoma Metastasis and Progression by Directly Targeting CDH6. Mol. Ther. 2018, 26, 1299–1312. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Yao, Q.; Hou, Y.; Xu, M.; Liu, S.; Yang, L.; Zhang, L.; Xu, H. MiR-223/Ect2/p21 signaling regulates osteosarcoma cell cycle progression and proliferation. Biomed. Pharmacother. 2019, 67, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.K.; Law, P.T.; Lee, C.W.; Cho, C.H.; Fan, D.; Wu, K.; Yu, J.; Sung, J.J. MicroRNA in colorectal cancer: From benchtop to bedside. Carcinogenesis 2010, 32, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhang, C.; Li, X.; Sun, W.; Qin, S.; Qin, L.; Wang, X. miR-223 promotes colon cancer by directly targeting p120 catenin. Oncotarget 2017, 8, 63764–63779. [Google Scholar] [CrossRef]
- Chai, B.; Guo, Y.; Cui, X.; Liu, J.; Suo, Y.; Dou, Z.; Li, N. MiR-223-3p promotes the proliferation, invasion and migration of colon cancer cells by negative regulating PRDM1. Am. J. Transl. Res. 2019, 11, 4516–4523. [Google Scholar]
- Bloomston, M.; Frankel, W.L.; Petrocca, F.; Volinia, S.; Alder, H.; Hagan, J.P.; Liu, C.-G.; Bhatt, D.; Taccioli, C.; Croce, C.M. MicroRNA Expression Patterns to Differentiate Pancreatic Adenocarcinoma from Normal Pancreas and Chronic Pancreatitis. JAMA 2019, 297, 1901–1908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fazi, F.; Racanicchi, S.; Zardo, G.; Starnes, L.; Mancini, M.; Travaglini, L.; Diverio, D.; Ammatuna, E.; Cimino, G.; Lo-Coco, F.; et al. Epigenetic Silencing of the Myelopoiesis Regulator microRNA-223 by the AML1/ETO Oncoprotein. Cancer Cell. 2007, 12, 457–466. [Google Scholar] [CrossRef]
- Gao, Y.; Lin, L.; Li, T.; Yang, J.; Wei, Y. The role of miRNA-223 in cancer: Function, diagnosis and therapy. Gene 2017, 616, 1–7. [Google Scholar] [CrossRef]
- Mavrakis, K.J.; Van Der Meulen, J.; Wolfe, A.L.; Liu, X.; Mets, E.; Taghon, T.; Khan, A.A.; Setty, M.; Rondou, P.; Vandenberghe, P.; et al. A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL). Nat. Genet. 2011, 43, 673–678. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.-Y.; Han, J.; Chen, X.-W.; Wang, J.; Wang, X.-D.; Sun, J.-G.; Chen, Z.-T. MiR-223 enhances the sensitivity of non-small cell lung cancer cells to erlotinib by targeting the insulin-like growth factor-1 receptor. Int. J. Mol. Med. 2016, 38, 183–191. [Google Scholar] [CrossRef] [Green Version]
- Tachibana, H.; Sho, R.; Takeda, Y.; Zhang, X.; Yoshida, Y.; Narimatsu, H.; Otani, K.; Ishikawa, S.; Fukao, A.; Asao, H.; et al. Circulating miR-223 in Oral Cancer: Its Potential as a Novel Diagnostic Biomarker and Therapeutic Target. PLoS ONE 2016, 11, e0159693. [Google Scholar] [CrossRef]
- Petrocca, F.; Visone, R.; Onelli, M.R.; Shah, M.H.; Nicoloso, M.; de Martino, I.; Iliopoulos, D.; Pilozzi, E.; Liu, C.-G.; Negrini, M.; et al. E2F1-Regulated MicroRNAs Impair TGFβ-Dependent Cell-Cycle Arrest and Apoptosis in Gastric Cancer. Cancer Cell. 2008, 13, 272–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Y.; Yang, J.; Yi, L.; Wang, Y.; Dong, Z.; Liu, Z.; Ou-Yang, S.; Wu, H.; Zhong, Z.; Yin, Z.; et al. MiR-223-3p targeting SEPT6 promotes the biological behavior of prostate cancer. Sci. Rep. 2014, 4, 7546. [Google Scholar] [CrossRef] [PubMed]
- Enagpal, N.; Kulshreshtha, R. MiR-191: An emerging player in disease biology. Front. Genet. 2014, 5, 99. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Li, H.; Jia, C.Y.; Cheng, W.; Yu, M.; Peng, M.; Zhu, Y.; Zhao, Q.; Dong, Y.W.; Shao, K.; et al. MicroRNA-223 regulates FOXO1 expression and cell proliferation. FEBS Lett. 2012, 586, 1038–1043. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.; Yan, C.; Liu, B.; Yang, C.; Nie, X.; Wang, X.; Zheng, J.; Wang, Y.; Zhu, Y. MiR-381 Regulates Neural Stem Cell Proliferation and Differentiation via Regulating Hes1 Expression. PLoS ONE 2015, 10, e0138973. [Google Scholar] [CrossRef]
- Najafi, Z.; Sharifi, M.; Javadi, G. Degradation of miR-21 induces apoptosis and inhibits cell proliferation in human hepatocellular carcinoma. Cancer Gene Ther. 2015, 22, 530–535. [Google Scholar] [CrossRef]
- Guz, M.; Rivero-Müller, A.; Okoń, E.; Stenzel-Bembenek, A.; Polberg, K.; Słomka, M.; Stepulak, A. MicroRNAs-Role in Lung Cancer. Dis. Markers. 2014, 2014, 218169. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Sengupta, T.; Kukreja, L.; Minella, A.C. MicroRNA-223 Regulates Cyclin E Activity by Modulating Expression of F-box and WD-40 Domain Protein 7. J. Biol. Chem. 2010, 285, 34439–34446. [Google Scholar] [CrossRef] [Green Version]
- Cells, A.T.L.; Moles, R.; Bellon, M.; Nicot, C. STAT1: A Novel Target of miR-150 and miR-223 Is Involved in the Proliferation of HTLV-I–Transformed. Neoplasia 2015, 17, 449–462. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Lan, X.; Li, D.; Li, T.; Lu, S. MiR-223 targeting MAFB suppresses proliferation and migration of nasopharyngeal carcinoma cells. BMC Cancer. 2015, 15, 461. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Guo, Y.; Liang, X.; Sun, M.; Wang, G.; De, W.; Wu, W. MicroRNA-223 functions as an oncogene in human gastric cancer by targeting FBXW7 / hCdc4. J Cancer Res Clin Oncol. 2012, 138, 763–774. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.-H.; Cheng, L.-H.; Hsu, H.-H.; Ho, T.-J.; Tu, C.-C.; Lin, Y.-M.; Chen, M.-C.; Tsai, F.-J.; Hsieh, Y.-L.; Huang, C.-Y. Apicidin-Resistant HA22T Hepatocellular Carcinoma Cells strongly activated the Wnt/β-Catenin Signaling Pathway and MMP-2 Expression via the IGF-IR/PI3K/Akt Signaling Pathway Enhancing Cell Metastatic Effect Apicidin-Resistant HA22T Hepatocellular. Biosci. Biotechnol. Biochem. 2013, 77, 2397–2404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Xu, L.; Li, C.; Zhao, L.; Ma, Y.; Zheng, H.; Li, Z.; Zhang, Y.; Wang, R.; Liu, Y.; et al. Ubiquitin ligase Cbl-b represses IGF-I-induced epithelial mesenchymal transition via ZEB2 and microRNA-200c regulation in gastric cancer cells. Mol. Cancer. 2014, 13, 136. [Google Scholar] [CrossRef] [Green Version]
- Jia, C.Y.; Li, H.H.; Zhu, X.C.; Dong, Y.W.; Fu, D.; Zhao, Q.L.; Wu, W.; Wu, X.Z. MiR-223 Suppresses Cell Proliferation by Targeting IGF-1R. PLoS ONE 2011, 6, e27008. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Ji, W.; Liu, X.; Ouyang, G.; Xiao, W. ELL Inhibits E2F1 Transcriptional Activity by Enhancing E2F1 Deacetylation via Recruitment of Histone Deacetylase 1. Mol. Cell. Biol. 2014, 34, 765–775. [Google Scholar] [CrossRef] [Green Version]
- Pulikkan, J.A.; Dengler, V.; Peramangalam, P.S.; Peer Zada, A.A.; Müller-Tidow, C.; Bohlander, S.K.; Tenen, D.G.; Behre, G. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood 2010, 115, 1768–1779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaddar, T.; Rouault, J.-P.; Chien, W.W.; Chebel, A.; Gadoux, M.; Salles, G.; Ffrench, M.; Magaud, J.-P. Two new miR-16 targets: Caprin-1 and HMGA1, proteins implicated in cell proliferation. Biol. Cell 2009, 101, 511–524. [Google Scholar] [CrossRef]
- Wang, B.; David, M.D.; Schrader, J.W. Absence of Caprin-1 Results in Defects in Cellular Proliferation. J. Immunol. 2005, 175, 4274–4282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Li, Z.; Guo, F.; Qin, X.; Liu, B.; Lei, Z.; Song, Z.; Sun, L.; Zhang, H.-T.; You, J.; et al. miR-223 regulates migration and invasion by targeting Artemin in human esophageal carcinoma. J. Biomed. Sci. 2011, 18, 24. [Google Scholar] [CrossRef] [Green Version]
- Li, B.-S.; Zhao, Y.-L.; Guo, G.; Li, W.; Zhu, E.-D.; Luo, X.; Mao, X.-H.; Zou, Q.-M.; Yu, P.-W.; Zuo, Q.-F.; et al. Plasma microRNAs, miR-223, miR-21 and miR-218, as Novel Potential Biomarkers for Gastric Cancer Detection. PLoS ONE 2012, 7, e41629. [Google Scholar] [CrossRef] [Green Version]
- Habbe, N.; Koorstra, J.-B.M.; Mendell, J.T.; Offerhaus, G.J.; Ryu, J.K.; Feldmann, G.; Mullendore, M.E.; Goggins, M.G.; Hong, S.-M.; Maitra, A. MicroRNA miR-155 is a biomarker of early pancreatic neoplasia. Cancer Biol. Ther. 2009, 8, 340–346. [Google Scholar] [CrossRef] [Green Version]
- Gomes, C.; Almeida, A.; Ferreira, J.A.; da Silva, M.L.S.; Santos-Sousa, H.; Pinto-De-Sousa, J.; Santos, L.L.; Amado, F.; Schwientek, T.; Levery, S.B.; et al. Glycoproteomic Analysis of Serum from Patients with Gastric Precancerous Lesions. J. Proteome Res. 2013, 12, 1454–1466. [Google Scholar] [CrossRef]
- Aziz, F.; Yang, X.; Wang, X.; Qiu, Y. Helicobacter Pylori infection leads to gastric cancer with the co-expression of Lewis Y and CA724. Glycoconj J. 2013, 30, 411. [Google Scholar]
- Chang, P.-Y.; Chen, C.-C.; Chang, Y.-S.; Tsai, W.-S.; You, J.-F.; Lin, G.-P.; Chen, T.; Chen, J.-S.; Chan, E.-C. MicroRNA-223 and microRNA-92a in stool and plasma samples act as complementary biomarkers to increase colorectal cancer detection. Oncotarget 2016, 7, 10663–10675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aziz, F.; Gao, W.; Yan, Q. Fucosyltransferase-4 and Oligosaccharide Lewis Y Antigen as potentially Correlative Biomarkers of Helicobacter pylori CagA Associated Gastric Cancer. Pathol. Oncol. Res. 2017, 23, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Aziz, F.; Wang, X.; Liu, J.; Yan, Q. Ginsenoside Rg3 induces FUT4-mediated apoptosis in H. pylori CagA-treated gastric cancer cells by regulating SP1 and HSF1 expressions. Toxicol In Vitro. 2016, 31, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Aziz, F.; Yang, X.; Wang, X.; Yan, Q. Anti-LeY antibody enhances therapeutic efficacy of celecoxib against gastric cancer by downregulation of MAPKs/COX-2 signaling pathway: Correlation with clinical study. J. Cancer Res. Clin. Oncol. 2015, 141, 1221–1235. [Google Scholar] [CrossRef] [PubMed]
- Aziz, F.; Chen, X.; Yang, X.; Yan, Q. Prevalence and Correlation with Clinical Diseases of Helicobacter pylori cagAandvacAGenotype among Gastric Patients from Northeast China. BioMed Res. Int. 2014, 2014, 142980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sibley, C.R.; Seow, Y.; Wood, M.J.A. Novel RNA-based Strategies for Therapeutic Gene Silencing. Mol. Ther. 2010, 18, 466–476. [Google Scholar] [CrossRef]
- Gao, C.; Cheng, X.; Li, X.; Tong, B.; Wu, K.; Liu, Y. Prognostic significance of artemin and GFRα1 expression in laryngeal squamous cell carcinoma. Exp. Ther. Med. 2014, 8, 818–822. [Google Scholar] [CrossRef]
- Escobar, G.; Gentner, B.; Naldini, L.; Mazzieri, R. Engineered tumor-infiltrating macrophages as gene delivery vehicles for interferon-α activates immunity and inhibits breast cancer progression. OncoImmunology 2014, 3, e28696. [Google Scholar] [CrossRef] [Green Version]
- Iliescu, L.; Mindrut, E.; Grasu, M.; Orban, C.; Tanase, A.; Toma, L. Management of Hepatocellular Carcinoma—Experience of a Single Center. Chirurgia 2014, 109, 204–207. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wu, C.; Che, X.; Wang, L.; Yu, D.; Zhang, T.; Huang, L.; Li, H.; Tan, W.; Wang, C.; et al. Circulating MicroRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol. Carcinog. 2011, 50, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Karakatsanis, A.; Papaconstantinou, I.; Gazouli, M.; Lyberopoulou, A.; Polymeneas, G.; Voros, D. Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol. Carcinog. 2013, 303, 297–303. [Google Scholar] [CrossRef]
- Pratedrat, P.; Chuaypen, N.; Nimsamer, P.; Payungporn, S.; Pinjaroen, N.; Sirichindakul, B.; Tangkijvanich, P. Diagnostic and prognostic roles of circulating miRNA-223-3p in hepatitis B virus–related hepatocellular carcinoma. PLoS ONE 2020, 15, e0232211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Jiang, Z.; Ma, N.; Wang, B.; Liu, J.; Zhang, L.; Gu, L. MicroRNA-223 Targeting STIM1 Inhibits the Biological Behavior of Breast Cancer. Cell. Physiol. Biochem. 2018, 45, 856–866. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhu, X.; Han, B.; Ji, L.; Yao, L.; Wang, Z. High Expression of microRNA-223 Indicates a Good Prognosis in Triple-Negative Breast Cancer. Front. Oncol. 2021, 11, 630432. [Google Scholar] [CrossRef] [PubMed]
- Su, X.-D.; Zhang, D.-K.; Zhang, X.; Lin, P.; Long, H.; Rong, T.-H. Prognostic factors in patients with recurrence after complete resection of esophageal squamous cell carcinoma. J. Thorac. Dis. 2014, 6, 949–957. [Google Scholar] [CrossRef] [PubMed]
- Ikeda-Miyagawa, Y.; Kobayashi, K.; Yamanaka, H.; Okubo, M.; Wang, S.; Dai, Y.; Yagi, H.; Hirose, M.; Noguchi, K. Peripherally Increased Artemin is a Key Regulator of TRPA1/V1 Expression in Primary Afferent Neurons. Mol. Pain. 2015, 11, 8–13. [Google Scholar] [CrossRef] [Green Version]
- Ding, K.; Banerjee, A.; Tan, S.; Zhao, J.; Zhuang, Q.; Li, R.; Qian, P.; Liu, S.; Wu, Z.-S.; Lobie, P.E.; et al. Artemin, a Member of the Glial Cell Line-derived Neurotrophic Factor Family of Ligands, Is HER2-regulated and Mediates Acquired Trastuzumab Resistance by Promoting Cancer Stem Cell-like Behavior in Mammary Carcinoma Cells. J. Biol. Chem. 2014, 289, 16057–16071. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.; Perry, J.K.; Pandey, V.; Fielder, G.C.; Mei, B.; Qian, P.X.; Wu, Z.S.; Zhu, T.; Liu, D.X.; Lobie, P.E. Artemin is oncogenic for human mammary carcinoma cells. Oncogen.e 2009, 28, 2034–2045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.; Li, M.; Hu, C.; Duan, H. Clinical significance of serum miR-223, miR-25 and miR-375 in patients with esophageal squamous cell carcinoma. Mol. Biol. Rep. 2014, 41, 1257–1266. [Google Scholar] [CrossRef] [PubMed]
- Khandwala, H.M.; McCutcheon, I.E.; Flyvbjerg, A.; Friend, K.E.; Osteosarcoma, A. The Effects of Insulin-Like Growth Factors on Tumorigenesis and Neoplastic Growth. Endocr. Rev. 2000, 21, 215–244. [Google Scholar] [CrossRef] [PubMed]
- D’Antona, P.; Cattoni, M.; Dominioni, L.; Poli, A.; Moretti, F.; Cinquetti, R.; Gini, E.; Daffrè, E.; Noonan, U.M.; Imperatori, A.; et al. Serum miR-223: A Validated Biomarker for Detection of Early-Stage Non–Small Cell Lung Cancer. Cancer Epidemiol. Biomark. Prev. 2019, 28, 1926–1933. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Clinical Relevance | Molecular Mechanism Involved or Function | Experimental Model | Specific Targets | References |
|---|---|---|---|---|
| Breast cancer | miR-233-3p mimics inhibited the NLRP3-dependent processes in cancer cells by suppressing the NLRP3 and downstream factors, including PYD and CARD domain-containing protein, IL-1β, and IL-18. | Breast cell lines: HMEC, MDA-MB231, MCF-7, and SKBR3 Xenograft mouse model | NLRP3, PYD, CARD, IL1β, IL18 | PMID: 30747211 |
| Hepatocellular carcinoma (HCC) | Checkpoint kinase 1, DNA methyltransferase 1, baculoviral IAP repeat-containing 5, kinesin family member 23, and collagen, type I, α1 target genes were considered the hub genes of miR-223-3p in HCC, which are significantly upregulated in HCC | Database: GEO database, TCGA database, Meta-analysis., Bioinformatics evaluation, Protein–protein interaction (PPI) network construction | Checkpoint kinase 1, DNA methyltransferase 1, baculoviral IAP repeat-containing 5, kinesin family member 23, and collagen, type I, α1. | PMID: 29207133 |
| Prostate Cancer | miR-223 inhibited the malignant behavior of prostate cancer cells while EYA3/c-Myc had the opposite effect. Moreover, circGNG4 enhanced the expression of EYA3/c-Myc by sponging miR-223 to promote the growth of prostate cancer tumors in vivo. | Prostate clinical samples: Prostate Cancer and adjacent normal tissues. Prostate cell lines: PC-3, LNCaP, VCaP, and DUL145, and the human normal prostatic epithelial cell line, RWPE-1 Xenograft mouse model | circGNG4, EYA transcriptional coactivator, phosphatase 3 (EYA3)/c-Myc | PMID: 34395419 |
| Esophageal squamous cell carcinoma (ESCC) | High expression level of miR-223 had a significant adverse impact on the survival of ESCC patients through repression of the function of FBXW7. | ESCC clinical samples: ESCC Primary ESCC tissue samples and human ESCC cell lines: TE1, TE4, TE6, TE8, TE9, TE10, TE14, and TE15. | FBXW7 | PMID: 22108521 |
| Gastric cancer | miR-223 induced by the transcription factor Twist, post-transcriptionally downregulates EPB41L3 expression by directly targeting its 3′-untranslated regions. | Gastric clinical samples: Gastric cancer and normal gastric tissues, Gastric cell lines: HEK293 cell, GPG29 cell, immortalized normal gastric mucosa GES cell, normal stomach fibroblastic cell NSFC, non-metastatic gastric cancer SUN-1, KATO-III, NUGC-3 cells, and metastatic gastric cancer XGC-9811L, AGS, and N87. | EPB41L3 | PMID: 21628394 |
| Hepatocellular carcinoma (HCC) | miR-223 functions as a tumor suppressor and plays a critical role in inhibiting the tumorigenesis and promoting the apoptosis of HCC through the mTOR signaling pathway by targeting Rab1. | Hepatocellular cell lines: Huh7, Hep3B, Bel-7402, and the HepG2 cell lines | Rab1, mTOR | PMID: 27998765 |
| Lung cancer | miR-223 targets IGF-1R, a transmembrane receptor tyrosine kinase related to lung cancer oncogenesis, tumor growth, and cancer cell survival. miR-223 overexpression leads to tumor suppression in lung cancer | IGF-1R, | PMID: 10857553 | |
| Liver cancer | miR-223-3p down-regulated the expression of FAT1, and inhibited the proliferation, migration, invasion, and EMT of liver cancer cells by targeting FAT1. FAT1 was highly expressed in liver cancer tissues and cells while miR-223-3p was lowly expressed. | Liver clinical samples: Liver Tumor tissues and corresponding adjacent tissues. Liver cell lines: Normal human hepatocyte, L-02 and human liver cancer cell lines, Hep G2, SMMC-7721, Hep 3b, and Li-7, | FAT1 | PMID: 32233593 |
| Brain cancer | miR-223-3p could be decreased by NLRP3 overexpression, which was considered as one of target genes of miR-223-3p. miR-223-3p might act as a suppressor and a potential therapy target of glioblastoma. | NLRP3 | PMID: 30033329 | |
| Colon cancer | miR-223 regulate the FoxO3a/BIM signaling pathway and colorectal cancer cell proliferation and apoptosis. Downregulating the expression of miR-223 increased the expression of FoxO3a and BIM and weakened cell proliferation and induced apoptosis. | Colon cell lines: NCM460 and SW620 cancer cells. | FoxO3 | PMID: 29949152 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aziz, F.; Chakraborty, A.; Khan, I.; Monts, J. Relevance of miR-223 as Potential Diagnostic and Prognostic Markers in Cancer. Biology 2022, 11, 249. https://doi.org/10.3390/biology11020249
Aziz F, Chakraborty A, Khan I, Monts J. Relevance of miR-223 as Potential Diagnostic and Prognostic Markers in Cancer. Biology. 2022; 11(2):249. https://doi.org/10.3390/biology11020249
Chicago/Turabian StyleAziz, Faisal, Abhijit Chakraborty, Imran Khan, and Josh Monts. 2022. "Relevance of miR-223 as Potential Diagnostic and Prognostic Markers in Cancer" Biology 11, no. 2: 249. https://doi.org/10.3390/biology11020249
APA StyleAziz, F., Chakraborty, A., Khan, I., & Monts, J. (2022). Relevance of miR-223 as Potential Diagnostic and Prognostic Markers in Cancer. Biology, 11(2), 249. https://doi.org/10.3390/biology11020249