Ex Vivo Rat Transected Spinal Cord Slices as a Model to Assess Lentiviral Vector Delivery of Neurotrophin-3 and Short Hairpin RNA against NG2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Dorsal Root Ganglia (DRG) Primary Cells
2.3. NT-3 and pWPT-GFP Lentiviral Vectors
2.4. shRNA Lentiviral Vectors Targeting NG2 Transcripts
2.5. Titration of Lentiviral Vectors
2.6. NT-3 ELISA
2.7. DRG Neurite Outgrowth Assay
2.8. Immunocytochemical Staining
2.8.1. DRG Immunocytochemical Staining
2.8.2. Neu7 Cell Immunocytochemical Staining
2.9. shNG2 Lentiviral Vector Transduction of Neu7 Cells
2.10. NG2 Western Blot
2.11. Measurement of Corrected Total Cell Fluorescence (CTCF) of NG2 in Transduced Neu7 cells
2.12. Animals
2.13. Spinal Cord Transection and Lentiviral Vector Transduction
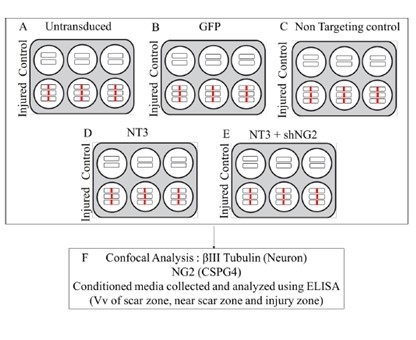
2.14. NT-3 and shNG2 Lentiviral Vector Transduction of Spinal Cord Slices
2.15. Tissue Fixation and Immunohistochemistry
2.16. Imaging
2.17. Stereology
2.18. Statistics
3. Results
3.1. Selection of shNG2 Lentiviral Vectors
3.2. Cells Transduced with NT-3 Lentiviral Vector Produce Significant Levels of NT-3 Protein Which Is Able to Promote DRG Neurite Outgrowth
3.3. Transduction with Lentiviral shNG2 Reduced NG2 Protein Neu7 Cells
3.4. Lentiviral shNG2 Knockdown of NG2 Promotes DRG Neurite Outgrowth In Vitro
3.5. Spinal Cord Slice Cultures Transduced with NT-3 Lentiviral Vector Produce NT-3 Protein
3.6. NG2 Expression Is Reduced in Ex Vivo Spinal Cord Slice Cultures 7 Days after Transduction with Lenti-NT-3/NG2 sh1
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alizadeh, A.; Dyck, S.M.; Karimi-Abdolrezaee, S. Traumatic Spinal Cord Injury: An Overview of Pathophysiology, Models and Acute Injury Mechanisms. Front. Neurol. 2019, 10, 282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lammertse, D.P. Clinical trials in spinal cord injury: Lessons learned on the path to translation. The 2011 International Spinal Cord Society Sir Ludwig Guttmann Lecture. Spinal Cord 2013, 51, 2–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharif-Alhoseini, M.; Khormali, M.; Rezaei, M.; Safdarian, M.; Hajighadery, A.; Khalatbari, M.M.; Safdarian, M.; Meknatkhah, S.; Rezvan, M.; Chalangari, M.; et al. Animal models of spinal cord injury: A systematic review. Spinal Cord 2017. [Google Scholar] [CrossRef] [PubMed]
- Pandamooz, S.; Nabiuni, M.; Miyan, J.; Ahmadiani, A.; Dargahi, L. Organotypic Spinal Cord Culture: A Proper Platform for the Functional Screening. Mol. Neurobiol. 2016, 53, 4659–4674. [Google Scholar] [CrossRef]
- Patar, A.; Dockery, P.; Howard, L.; McMahon, S. Analysis of reactive astrocytes and NG2 proteoglycan in ex vivo rat models of spinal cord injury. J. Neurosci. Methods 2019, 311, 418–425. [Google Scholar] [CrossRef] [Green Version]
- Ravikumar, M.; Jain, S.; Miller, R.H.; Capadona, J.R.; Selkirk, S.M. An organotypic spinal cord slice culture model to quantify neurodegeneration. J. Neurosci. Methods 2012, 211, 280–288. [Google Scholar] [CrossRef]
- Morrison, B., 3rd; Saatman, K.E.; Meaney, D.F.; McIntosh, T.K. In vitro central nervous system models of mechanically induced trauma: A review. J. Neurotrauma 1998, 15, 911–928. [Google Scholar] [CrossRef] [Green Version]
- Sypecka, J.; Koniusz, S.; Kawalec, M.; Sarnowska, A. The organotypic longitudinal spinal cord slice culture for stem cell study. Stem Cells Int. 2015, 2015, 471216. [Google Scholar] [CrossRef]
- Pohland, M.; Glumm, R.; Stoenica, L.; Holtje, M.; Kiwit, J.; Ahnert-Hilger, G.; Strauss, U.; Brauer, A.U.; Paul, F.; Glumm, J. Studying axonal outgrowth and regeneration of the corticospinal tract in organotypic slice cultures. J. Neurotrauma 2015. [Google Scholar] [CrossRef]
- Weightman, A.P.; Pickard, M.R.; Yang, Y.; Chari, D.M. An in vitro spinal cord injury model to screen neuroregenerative materials. Biomaterials 2014, 35, 3756–3765. [Google Scholar] [CrossRef] [Green Version]
- Pinkernelle, J.; Fansa, H.; Ebmeyer, U.; Keilhoff, G. Prolonged minocycline treatment impairs motor neuronal survival and glial function in organotypic rat spinal cord cultures. PLoS ONE 2013, 8, e73422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krassioukov, A.V.; Ackery, A.; Schwartz, G.; Adamchik, Y.; Liu, Y.; Fehlings, M.G. An in vitro model of neurotrauma in organotypic spinal cord cultures from adult mice. Brain Res. Brain Res. Protoc. 2002, 10, 60–68. [Google Scholar] [CrossRef]
- Cho, J.S.; Park, H.W.; Park, S.K.; Roh, S.; Kang, S.K.; Paik, K.S.; Chang, M.S. Transplantation of mesenchymal stem cells enhances axonal outgrowth and cell survival in an organotypic spinal cord slice culture. Neurosci. Lett. 2009, 454, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Naldini, L.; Blomer, U.; Gallay, P.; Ory, D.; Mulligan, R.; Gage, F.H.; Verma, I.M.; Trono, D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 1996, 272, 263–267. [Google Scholar] [CrossRef] [Green Version]
- Blomer, U.; Naldini, L.; Kafri, T.; Trono, D.; Verma, I.M.; Gage, F.H. Highly efficient and sustained gene transfer in adult neurons with a lentivirus vector. J. Virol. 1997, 71, 6641–6649. [Google Scholar] [CrossRef] [Green Version]
- Hutson, T.H.; Foster, E.; Moon, L.D.; Yanez-Munoz, R.J. Lentiviral vector-mediated RNA silencing in the central nervous system. Hum. Gene Ther. Methods 2014, 25, 14–32. [Google Scholar] [CrossRef] [Green Version]
- Sakuma, T.; Barry, M.A.; Ikeda, Y. Lentiviral vectors: Basic to translational. Biochem. J. 2012, 443, 603–618. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Jiang, X.; Zhang, L.; Pu, H.; Hu, X.; Zhang, W.; Cai, W.; Gao, Y.; Leak, R.K.; Keep, R.F.; et al. Endothelium-targeted overexpression of heat shock protein 27 ameliorates blood-brain barrier disruption after ischemic brain injury. Proc. Natl. Acad. Sci. USA 2017, 114, E1243–E1252. [Google Scholar] [CrossRef] [Green Version]
- Kelly, M.J.; O’Keeffe, G.W.; Sullivan, A.M. Viral vector delivery of neurotrophic factors for Parkinson’s disease therapy. Expert Rev. Mol. Med. 2015, 17, e8. [Google Scholar] [CrossRef]
- Taylor, L.; Jones, L.; Tuszynski, M.H.; Blesch, A. Neurotrophin-3 gradients established by lentiviral gene delivery promote short-distance axonal bridging beyond cellular grafts in the injured spinal cord. J. Neurosci. 2006, 26, 9713–9721. [Google Scholar] [CrossRef] [Green Version]
- Bonner, J.F.; Blesch, A.; Neuhuber, B.; Fischer, I. Promoting directional axon growth from neural progenitors grafted into the injured spinal cord. J. Neurosci. Res. 2010, 88, 1182–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, R.R.; Muir, E.M.; Alves, J.N.; Rickman, H.; Allan, A.Y.; Kwok, J.C.; Roet, K.C.; Verhaagen, J.; Schneider, B.L.; Bensadoun, J.C.; et al. Lentiviral vectors express chondroitinase ABC in cortical projections and promote sprouting of injured corticospinal axons. J. Neurosci. Methods 2011, 201, 228–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartus, K.; James, N.D.; Didangelos, A.; Bosch, K.D.; Verhaagen, J.; Yáñez-Muñoz, R.J.; Rogers, J.H.; Schneider, B.L.; Muir, E.M.; Bradbury, E.J. Large-Scale Chondroitin Sulfate Proteoglycan Digestion with Chondroitinase Gene Therapy Leads to Reduced Pathology and Modulates Macrophage Phenotype following Spinal Cord Contusion Injury. J. Neurosci. 2014, 34, 4822–4836. [Google Scholar] [CrossRef] [PubMed]
- Kusano, K.; Enomoto, M.; Hirai, T.; Tsoulfas, P.; Sotome, S.; Shinomiya, K.; Okawa, A. Transplanted neural progenitor cells expressing mutant NT3 promote myelination and partial hindlimb recovery in the chronic phase after spinal cord injury. Biochem. Biophys. Res. Commun. 2010, 393, 812–817. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, E.J.; Moon, L.D.; Popat, R.J.; King, V.R.; Bennett, G.S.; Patel, P.N.; Fawcett, J.W.; McMahon, S.B. Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature 2002, 416, 636–640. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.; Nicholson, L.; van Niekerk, E.; Motsch, M.; Blesch, A. DEPENDENCE OF REGENERATED SENSORY AXONS ON CONTINUOUS NEUROTROPHIN-3 DELIVERY. J. Neurosci. Off. J. Soc. Neurosci. 2012, 32, 13206–13220. [Google Scholar] [CrossRef] [Green Version]
- Lynskey, J.V.; Sandhu, F.A.; Dai, H.N.; McAtee, M.; Slotkin, J.R.; MacArthur, L.; Bregman, B.S. Delayed intervention with transplants and neurotrophic factors supports recovery of forelimb function after cervical spinal cord injury in adult rats. J. Neurotrauma 2006, 23, 617–634. [Google Scholar] [CrossRef]
- Hwang, D.H.; Kim, H.M.; Kang, Y.M.; Joo, I.S.; Cho, C.S.; Yoon, B.W.; Kim, S.U.; Kim, B.G. Combination of multifaceted strategies to maximize the therapeutic benefits of neural stem cell transplantation for spinal cord repair. Cell Transpl. 2011, 20, 1361–1379. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Zhang, A.; Duan, H.; Zhang, S.; Hao, P.; Ye, K.; Sun, Y.E.; Li, X. NT3-chitosan elicits robust endogenous neurogenesis to enable functional recovery after spinal cord injury. Proc. Natl. Acad. Sci. USA 2015, 112, 13354–13359. [Google Scholar] [CrossRef] [Green Version]
- Ziemba, A.M.; Gilbert, R.J. Biomaterials for Local, Controlled Drug Delivery to the Injured Spinal Cord. Front. Pharmacol. 2017, 8, 245. [Google Scholar] [CrossRef]
- McMahon, S.S.; Nikolskaya, N.; Choileain, S.N.; Hennessy, N.; O’Brien, T.; Strappe, P.M.; Gorelov, A.; Rochev, Y. Thermosensitive hydrogel for prolonged delivery of lentiviral vector expressing neurotrophin-3 in vitro. J. Gene Med. 2011, 13, 591–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alto, L.T.; Havton, L.A.; Conner, J.M.; Hollis, E.R., 2nd; Blesch, A.; Tuszynski, M.H. Chemotropic guidance facilitates axonal regeneration and synapse formation after spinal cord injury. Nat. Neurosci. 2009, 12, 1106–1113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desclaux, M.; Teigell, M.; Amar, L.; Vogel, R.; Gimenez, Y.R.M.; Privat, A.; Mallet, J. A novel and efficient gene transfer strategy reduces glial reactivity and improves neuronal survival and axonal growth in vitro. PLoS ONE 2009, 4, e6227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desclaux, M.; Perrin, F.E.; Do-Thi, A.; Prieto-Cappellini, M.; Gimenez, Y.R.M.; Mallet, J.; Privat, A. Lentiviral-mediated silencing of glial fibrillary acidic protein and vimentin promotes anatomical plasticity and functional recovery after spinal cord injury. J. Neurosci. Res. 2015, 93, 43–55. [Google Scholar] [CrossRef]
- Donnelly, E.M.; Madigan, N.N.; Rooney, G.E.; Knight, A.; Chen, B.; Ball, B.; Kinnavane, L.; Garcia, Y.; Dockery, P.; Fraher, J.; et al. Lentiviral vector delivery of short hairpin RNA to NG2 and neurotrophin-3 promotes locomotor recovery in injured rat spinal cord. Cytotherapy 2012, 14, 1235–1244. [Google Scholar] [CrossRef]
- Donnelly, E.M.; Strappe, P.M.; McGinley, L.M.; Madigan, N.N.; Geurts, E.; Rooney, G.E.; Windebank, A.J.; Fraher, J.; Dockery, P.; O’Brien, T.; et al. Lentiviral vector-mediated knockdown of the NG2 [corrected] proteoglycan or expression of neurotrophin-3 promotes neurite outgrowth in a cell culture model of the glial scar. J. Gene Med. 2010, 12, 863–872. [Google Scholar] [CrossRef]
- Zhou, H.X.; Li, X.Y.; Li, F.Y.; Liu, C.; Liang, Z.P.; Liu, S.; Zhang, B.; Wang, T.Y.; Chu, T.C.; Lu, L.; et al. Targeting RPTPsigma with lentiviral shRNA promotes neurites outgrowth of cortical neurons and improves functional recovery in a rat spinal cord contusion model. Brain Res. 2014, 1586, 46–63. [Google Scholar] [CrossRef]
- Sandvig, A.; Berry, M.; Barrett, L.B.; Butt, A.; Logan, A. Myelin-, reactive glia-, and scar-derived CNS axon growth inhibitors: Expression, receptor signaling, and correlation with axon regeneration. Glia 2004, 46, 225–251. [Google Scholar] [CrossRef]
- Ampofo, E.; Schmitt, B.M.; Menger, M.D.; Laschke, M.W. The regulatory mechanisms of NG2/CSPG4 expression. Cell Mol. Biol. Lett. 2017, 22, 4. [Google Scholar] [CrossRef] [Green Version]
- Levine, J. The reactions and role of NG2 glia in spinal cord injury. Brain Res. 2016, 1638, 199–208. [Google Scholar] [CrossRef] [Green Version]
- Greenwood, K.; Butt, A.M. Evidence that perinatal and adult NG2-glia are not conventional oligodendrocyte progenitors and do not depend on axons for their survival. Mol. Cell Neurosci. 2003, 23, 544–558. [Google Scholar] [CrossRef]
- Hackett, A.R.; Lee, J.K. Understanding the NG2 Glial Scar after Spinal Cord Injury. Front. Neurol. 2016, 7, 199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- James, N.D.; Shea, J.; Muir, E.M.; Verhaagen, J.; Schneider, B.L.; Bradbury, E.J. Chondroitinase gene therapy improves upper limb function following cervical contusion injury. Exp. Neurol. 2015, 271, 131–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwok, J.C.; Yuen, Y.L.; Lau, W.K.; Zhang, F.X.; Fawcett, J.W.; Chan, Y.S.; Shum, D.K. Chondroitin sulfates in the developing rat hindbrain confine commissural projections of vestibular nuclear neurons. Neural Dev. 2012, 7, 6. [Google Scholar] [CrossRef] [Green Version]
- Moon, L.D.; Asher, R.A.; Rhodes, K.E.; Fawcett, J.W. Regeneration of CNS axons back to their target following treatment of adult rat brain with chondroitinase ABC. Nat. Neurosci. 2001, 4, 465–466. [Google Scholar] [CrossRef]
- Kilcoyne, M.; Sharma, S.; McDevitt, N.; O’Leary, C.; Joshi, L.; McMahon, S.S. Neuronal glycosylation differentials in normal, injured and chondroitinase-treated environments. Biochem. Biophys. Res. Commun. 2012, 420, 616–622. [Google Scholar] [CrossRef] [Green Version]
- Fok-Seang, J.; Smith-Thomas, L.C.; Meiners, S.; Muir, E.; Du, J.S.; Housden, E.; Johnson, A.R.; Faissner, A.; Geller, H.M.; Keynes, R.J.; et al. An analysis of astrocytic cell lines with different abilities to promote axon growth. Brain Res. 1995, 689, 207–223. [Google Scholar] [CrossRef]
- Levin, E.; Diekmann, H.; Fischer, D. Highly efficient transduction of primary adult CNS and PNS neurons. Sci. Rep. 2016, 6, 38928. [Google Scholar] [CrossRef] [Green Version]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef] [Green Version]
- Kutner, R.H.; Zhang, X.Y.; Reiser, J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat. Protoc. 2009, 4, 495–505. [Google Scholar] [CrossRef]
- Sastry, L.; Johnson, T.; Hobson, M.J.; Smucker, B.; Cornetta, K. Titering lentiviral vectors: Comparison of DNA, RNA and marker expression methods. Gene 2002, 9, 1155–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennedy, H.S.; Jones, C., 3rd; Caplazi, P. Comparison of standard laminectomy with an optimized ejection method for the removal of spinal cords from rats and mice. J. Histotechnol. 2013, 36, 86–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patar, A.; Dockery, P.; Howard, L.; McMahon, S.S. Cell viability in three ex vivo rat models of spinal cord injury. J. Anat. 2019, 234, 244–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith-Thomas, L.C.; Fok-Seang, J.; Stevens, J.; Du, J.S.; Muir, E.; Faissner, A.; Geller, H.M.; Rogers, J.H.; Fawcett, J.W. An inhibitor of neurite outgrowth produced by astrocytes. J. Cell Sci 1994, 107 Pt 6, 1687–1695. [Google Scholar]
- Fidler, P.S.; Schuette, K.; Asher, R.A.; Dobbertin, A.; Thornton, S.R.; Calle-Patino, Y.; Muir, E.; Levine, J.M.; Geller, H.M.; Rogers, J.H.; et al. Comparing astrocytic cell lines that are inhibitory or permissive for axon growth: The major axon-inhibitory proteoglycan is NG2. J. Neurosci. 1999, 19, 8778–8788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laabs, T.L.; Wang, H.; Katagiri, Y.; McCann, T.; Fawcett, J.W.; Geller, H.M. Inhibiting glycosaminoglycan chain polymerization decreases the inhibitory activity of astrocyte-derived chondroitin sulfate proteoglycans. J. Neurosci. 2007, 27, 14494–14501. [Google Scholar] [CrossRef]
- Kumar, S.; Biancotti, J.C.; Yamaguchi, M.; de Vellis, J. Combination of growth factors enhances remyelination in a cuprizone-induced demyelination mouse model. Neurochem. Res. 2007, 32, 783–797. [Google Scholar] [CrossRef]
- Hodgetts, S.I.; Harvey, A.R. Neurotrophic Factors Used to Treat Spinal Cord Injury. Vitam. Horm. 2017, 104, 405–457. [Google Scholar] [CrossRef]
- Keefe, K.M.; Sheikh, I.S.; Smith, G.M. Targeting Neurotrophins to Specific Populations of Neurons: NGF, BDNF, and NT-3 and Their Relevance for Treatment of Spinal Cord Injury. Int. J. Mol. Sci. 2017, 18, 548. [Google Scholar] [CrossRef]
- Hutson, T.H.; Foster, E.; Dawes, J.M.; Hindges, R.; Yanez-Munoz, R.J.; Moon, L.D. Lentiviral vectors encoding short hairpin RNAs efficiently transduce and knockdown LINGO-1 but induce an interferon response and cytotoxicity in central nervous system neurones. J. Gene Med. 2012, 14, 299–315. [Google Scholar] [CrossRef]
- Liu, G.M.; Luo, Y.G.; Li, J.; Xu, K. Knockdown of Nogo gene by short hairpin RNA interference promotes functional recovery of spinal cord injury in a rat model. Mol. Med. Rep. 2016, 13, 4431–4436. [Google Scholar] [CrossRef] [PubMed]
- Bonnici, B.; Kapfhammer, J.P. Spontaneous regeneration of intrinsic spinal cord axons in a novel spinal cord slice culture model. Eur. J. Neurosci. 2008, 27, 2483–2492. [Google Scholar] [CrossRef] [PubMed]
- Barrett, C.P.; Donati, E.J.; Guth, L. Differences between adult and neonatal rats in their astroglial response to spinal injury. Exp. Neurol. 1984, 84, 374–385. [Google Scholar] [CrossRef]










| ShRNAs | Species | DNA Sequences Targeting CSPG4 (NG2) | Alignment |
|---|---|---|---|
| NG2 sh1 | Rattus norvegicus | …GGGACAAGCGTGGCAACTTTATCTA… | Match |
| Catalogue number TRCN0000348358 | Mus musculus | …GGGACAAGCGTGGCAACTTTATCTA… | |
| NG2 sh2 | Rattus norvegicus | …GCATAGAAGATTTCAGTGTTAATGG… | Match |
| Catalogue number TRCN0000098337 | Mus musculus | …GCATAGAAGATTTCAGTGTTAATGG… | |
| NG2 sh3 | Rattus norvegicus | …GGGTATCTCCACGTAGCCAATAGT… | One base mismatch |
| Catalogue number TRCN0000334788 | Mus musculus | …CCCTATCTTCACGTAGCCAATAGT… | |
| NG2 sh4 | Rattus norvegicus | …CAATACCCTACACGTACTTTCAACC… | One base mismatch |
| Catalogue number TRCN0000348354 | Mus musculus | …CAATACCCTACGCGTACTTTCAACC… | |
| NG2 sh5 | Rattus norvegicus | …GCAACCAACTTGTGGAAGATTTC… | One base mismatch |
| Catalogue number TRCN0000098338 | Mus musculus | …GCAACCAACTTGTGGAACATTTC… |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patar, A.; Dockery, P.; McMahon, S.; Howard, L. Ex Vivo Rat Transected Spinal Cord Slices as a Model to Assess Lentiviral Vector Delivery of Neurotrophin-3 and Short Hairpin RNA against NG2. Biology 2020, 9, 54. https://doi.org/10.3390/biology9030054
Patar A, Dockery P, McMahon S, Howard L. Ex Vivo Rat Transected Spinal Cord Slices as a Model to Assess Lentiviral Vector Delivery of Neurotrophin-3 and Short Hairpin RNA against NG2. Biology. 2020; 9(3):54. https://doi.org/10.3390/biology9030054
Chicago/Turabian StylePatar, Azim, Peter Dockery, Siobhan McMahon, and Linda Howard. 2020. "Ex Vivo Rat Transected Spinal Cord Slices as a Model to Assess Lentiviral Vector Delivery of Neurotrophin-3 and Short Hairpin RNA against NG2" Biology 9, no. 3: 54. https://doi.org/10.3390/biology9030054
APA StylePatar, A., Dockery, P., McMahon, S., & Howard, L. (2020). Ex Vivo Rat Transected Spinal Cord Slices as a Model to Assess Lentiviral Vector Delivery of Neurotrophin-3 and Short Hairpin RNA against NG2. Biology, 9(3), 54. https://doi.org/10.3390/biology9030054