Mutually Exclusive Expression of Closely Related Odorant-Binding Proteins 9A and 9B in the Antenna of the Red Flour Beetle Tribolium castaneum
Abstract
:1. Introduction
2. Material and Methods
2.1. Tribolium Rearing
2.2. Cloning of TcasOBP9A and TcasOBP9B
2.3. RNA Interference
2.4. Electroantennography
2.5. Antennal Fluorescent In Situ Hybridization
2.6. Microscopy and Image Processing
2.7. Phylogenetic Analysis and Interspecies Comparison
2.8. Homology Modeling and In Silico Docking Experiments
3. Results and Discussion
3.1. TcasOBP9A and TcasOBP9B Enhance Detection of a Broad Spectrum of Volatiles
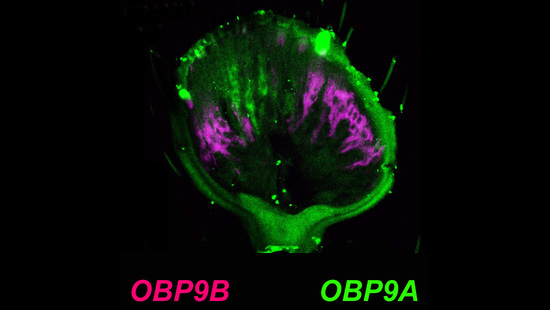
3.2. Mutual Exclusive Antennal Expression of TcasOBP9A and TcasOBP9B
3.3. Phylogeny of TcasOBP9A and TcasOBP9B
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leal, W.S. Odorant Reception in Insects: Roles of Receptors, Binding Proteins, and Degrading Enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef]
- Steinbrecht, R.A. Odorant-Binding Proteins: Expression and Function. Proc. Natl. Acad. Sci. USA 1998, 855, 323–332. [Google Scholar] [CrossRef]
- Vogt, R.G.; Riddiford, L.M. Pheromone binding and inactivation by moth antennae. Nature 1981, 293, 161–163. [Google Scholar] [CrossRef]
- Zhou, J.-J. Chapter Ten—Odorant-Binding Proteins in Insects. In Vitamins & Hormones Vol. 83, Pheromones; Litwack, G., Ed.; Academic Press: San Diego, CA, USA, 2010; pp. 241–272. [Google Scholar]
- Sandler, B.H.; Nikonova, L.; Leal, W.S.; Clardy, J. Sexual attraction in the silkworm moth: Structure of the pheromone-binding-protein-bombykol complex. Chem. Biol. 2000, 7, 143–151. [Google Scholar] [CrossRef] [Green Version]
- Swarup, S.; Williams, T.I.; Anholt, R.R.H. Functional dissection of Odorant binding protein genes in Drosophila melanogaster. Genes Brain Behav. 2011, 10, 648–657. [Google Scholar] [CrossRef]
- Xu, P.; Atkinson, R.; Jones, D.N.; Smith, D.P. Drosophila OBP LUSH Is Required for Activity of Pheromone-Sensitive Neurons. Neuron 2005, 45, 193–200. [Google Scholar] [CrossRef] [Green Version]
- Arya, G.H.; Weber, A.L.; Wang, P.; Magwire, M.M.; Negron, Y.L.S.; Mackay, T.F.C.; Anholt, R.R.H. Natural Variation, Functional Pleiotropy and Transcriptional Contexts of Odorant Binding Protein Genes in Drosophila melanogaster. Genetics 2010, 186, 1475–1485. [Google Scholar] [CrossRef] [Green Version]
- Krieger, M.J.B.; Ross, K.G. Molecular evolutionary analyses of the odorant-binding protein gene Gp-9 in fire ants and other Solenopsis species. Mol. Biol. Evol. 2005, 22, 2090–2103. [Google Scholar] [CrossRef] [Green Version]
- Biessmann, H.; Andronopoulou, E.; Biessmann, M.R.; Douris, V.; Dimitratos, S.D.; Eliopoulos, E.; Guerin, P.M.; Iatrou, K.; Justice, R.W.; Kröber, T. The Anopheles gambiae Odorant Binding Protein 1 (AgamOBP1) Mediates Indole Recognition in the Antennae of Female Mosquitoes. PLoS ONE 2010, 5, e9471. [Google Scholar] [CrossRef] [Green Version]
- Pelletier, J.; Guidolin, A.S.; Syed, Z.; Cornel, A.J.; Leal, W.S. Knockdown of a Mosquito Odorant-binding Protein Involved in the Sensitive Detection of Oviposition Attractants. J. Chem. Ecol. 2010, 36, 245–248. [Google Scholar] [CrossRef] [Green Version]
- Forstner, M. A receptor and binding protein interplay in the detection of a distinct pheromone component in the silkmoth Antheraea polyphemus. Int. J. Biol. Sci. 2009, 5, 745–757. [Google Scholar] [CrossRef] [Green Version]
- Grosse-Wilde, E.; Svatos, A.; Krieger, J. A pheromone-binding protein mediates the bombykol-induced activation of a pheromone receptor in vitro. Chem. Senses 2006, 31, 547–555. [Google Scholar] [CrossRef]
- Grosse-Wilde, E.; Gohl, T.; Bouché, E.; Breer, H.; Krieger, J. Candidate pheromone receptors provide the basis for the response of distinct antennal neurons to pheromonal compounds. Eur. J. Neurosci. 2007, 25, 2364–2373. [Google Scholar] [CrossRef]
- Hallem, E.A.; Ho, M.G.; Carlson, J.R. The molecular basis of odor coding in the Drosophila antenna. Cell 2004, 117, 965–979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Syed, Z.; Ishida, Y.; Taylor, K.; Kimbrell, D.A.; Leal, W.S. Pheromone reception in fruit flies expressing a moth’s odorant receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 16538–16543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziegelberger, G. Redox-Shift of the Pheromone-Binding Protein in the Silkmoth Antheraea Polyphemus. JBIC J. Biol. Inorg. Chem. 1995, 232, 706–711. [Google Scholar] [CrossRef]
- Kaissling, K.-E. Chemo-electrical transduction in insect olfactory receptors. Annu. Rev. Neurosci. 1986, 9, 121–145. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Sun, J.S.; Carlson, J.R. Robust olfactory responses in the absence of odorant binding proteins. eLife 2019, 8, e51040. [Google Scholar] [CrossRef]
- Larter, N.K.; Sun, J.S.; Carlson, J.R. Organization and function of Drosophila odorant binding proteins. eLife 2016, 5. [Google Scholar] [CrossRef]
- Dippel, S.; Oberhofer, G.; Kahnt, J.; Gerischer, L.; Opitz, L.; Schachtner, J.; Stanke, M.; Schütz, S.; Wimmer, E.A.; Angeli, S. Tissue-specific transcriptomics, chromosomal localization, and phylogeny of chemosensory and odorant binding proteins from the red flour beetle Tribolium castaneum reveal subgroup specificities for olfaction or more general functions. BMC Genom. 2014, 15, 1141. [Google Scholar] [CrossRef] [Green Version]
- Qiao, H.; He, X.; Schymura, D.; Ban, L.; Field, L.; Dani, F.R.; Michelucci, E.; Caputo, B.; della Torre, A.; Iatrou, K.; et al. Cooperative interactions between odorant-binding proteins of Anopheles gambiae. Cell. Mol. Life Sci. 2010, 68, 1799–1813. [Google Scholar] [CrossRef] [PubMed]
- Schultze, A.; Pregitzer, P.; Walter, M.F.; Woods, D.F.; Marinotti, O.; Breer, H.; Krieger, J. The Co-Expression Pattern of Odorant Binding Proteins and Olfactory Receptors Identify Distinct Trichoid Sensilla on the Antenna of the Malaria Mosquito Anopheles gambiae. PLoS ONE 2013, 8, e69412. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Diaz, C.; Reina, J.H.; Cambillau, C.; Benton, R. Ligands for Pheromone-Sensing Neurons Are Not Conformationally Activated Odorant Binding Proteins. PLoS Biol. 2013, 11, e1001546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kruse, S.W.; Zhao, R.; Smith, D.P.; Jones, D.N.M. Structure of a specific alcohol-binding site defined by the odorant binding protein LUSH from Drosophila melanogaster. Nat. Struct. Mol. Biol. 2003, 10, 694–700. [Google Scholar] [CrossRef] [Green Version]
- Laughlin, J.D.; Ha, T.S.; Jones, D.N.M.; Smith, D.P. Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell 2008, 133, 1255–1265. [Google Scholar] [CrossRef] [Green Version]
- Katti, S.; Lokhande, N.; González, D.; Cassill, A.; Renthal, R. Quantitative analysis of pheromone-binding protein specificity. Insect Mol. Biol. 2013, 22, 31–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bucci, B.K.; Kruse, S.W.; Thode, A.B.; Alvarado, S.M.; Jones, D.N.M. Effect of n-alcohols on the structure and stability of the Drosophila odorant binding protein LUSH. Biochemistry 2006, 45, 1693–1701. [Google Scholar] [CrossRef] [PubMed]
- Thode, A.B.; Kruse, S.W.; Nix, J.C.; Jones, D.N. The Role of Multiple Hydrogen-Bonding Groups in Specific Alcohol Binding Sites in Proteins: Insights from Structural Studies of LUSH. J. Mol. Biol. 2008, 376, 1360–1376. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.-J.; Zhang, G.-A.; Huang, W.; A Birkett, M.; Field, L.M.; Pickett, J.A.; Pelosi, P. Revisiting the odorant-binding protein LUSH ofDrosophila melanogaster: Evidence for odour recognition and discrimination. FEBS Lett. 2004, 558, 23–26. [Google Scholar] [CrossRef] [Green Version]
- Sokoloff, A. The Genetics of Tribolium and Related Species; Academic Press: New York, NY, USA, 1966. [Google Scholar]
- Brown, S.J.; Shippy, T.D.; Miller, S.; Bolognesi, R.; Beeman, R.W.; Lorenzen, M.D.; Bucher, G.; Wimmer, E.A.; Klingler, M. The red flour beetle, Tribolium castaneum (Coleoptera): A model for studies of development and pest biology. Cold Spring Harb. Protoc. 2009, 2009. [Google Scholar] [CrossRef]
- Bucher, G.; Scholten, J.; Klingler, M. Parental RNAi in Tribolium (Coleoptera). Curr. Biol. CB 2002, 12, R85–R86. [Google Scholar] [CrossRef] [Green Version]
- Tomoyasu, Y.; Denell, R.E. Larval RNAi in Tribolium (Coleoptera) for analyzing adult development. Dev. Genes Evol. 2004, 214, 575–578. [Google Scholar] [CrossRef] [PubMed]
- Trauner, J.; Schinko, J.; Lorenzen, M.D.; Shippy, T.D.; Wimmer, E.A.; Beeman, R.W.; Klingler, M.; Bucher, G.; Brown, S.J. Large-scale insertional mutagenesis of a coleopteran stored grain pest, the red flour beetle Tribolium castaneum, identifies embryonic lethal mutations and enhancer traps. BMC Biol. 2009, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilles, A.F.; Schinko, J.B.; Averof, M. Efficient CRISPR-mediated gene targeting and transgene replacement in the beetle Tribolium castaneum. Dev. Camb. Engl. 2015, 142, 2832–2839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schinko, J.B.; Weber, M.; Viktorinova, I.; Kiupakis, A.; Averof, M.; Klingler, M.; Wimmer, E.A.; Bucher, G. Functionality of the GAL4/UAS system in Tribolium requires the use of endogenous core promoters. BMC Dev. Biol. 2010, 10, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schinko, J.B.; Hillebrand, K.; Bucher, G. Heat shock-mediated misexpression of genes in the beetle Tribolium castaneum. Dev. Genes Evol. 2012, 222, 287–298. [Google Scholar] [CrossRef]
- Richards, S.; Gibbs, R.A.; Weinstock, G.M.; Brown, S.J.; Denell, R.; Beeman, R.W.; Gibbs, R.; Beeman, R.W.; Brown, S.J.; Bucher, G.; et al. The genome of the model beetle and pest Tribolium castaneum. Nature 2018, 452, 949–955. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.S.; Murphy, T.; Xia, J.; Caragea, D.; Park, Y.; Beeman, R.W.; Lorenzen, M.D.; Butcher, S.; Manak, J.R.; Brown, S.J. BeetleBase in 2010: Revisions to provide comprehensive genomic information for Tribolium castaneum. Nucleic Acids Res. 2010, 38, D437–D442. [Google Scholar] [CrossRef] [Green Version]
- Herndon, N.; Shelton, J.; Gerischer, L.; Ioannidis, P.; Ninova, M.; Dönitz, J.; Waterhouse, R.M.; Liang, C.; Damm, C.; Siemanowski, J.; et al. Enhanced genome assembly and a new official gene set for Tribolium castaneum. BMC Genom. 2020, 21, 47. [Google Scholar] [CrossRef] [Green Version]
- Balakrishnan, K.; Holighaus, G.; Weißbecker, B.; Schütz, S. Electroantennographic responses of red flour beetle Tribolium castaneum Herbst (Coleoptera: Tenebrionidae) to volatile organic compounds. J. Appl. Entomol. 2017, 141, 477–486. [Google Scholar] [CrossRef]
- Trebels, B.; Dippel, S.; Schaaf, M.; Balakrishnan, K.; Wimmer, E.A.; Schachtner, J. Adult neurogenesis in the mushroom bodies of red flour beetles (Tribolium castaneum, Herbst) is influenced by the olfactory environment. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Karner, T.; Kellner, I.; Schultze, A.; Breer, H.; Krieger, J. Co-expression of six tightly clustered odorant receptor genes in the antenna of the malaria mosquito Anopheles gambiae. Chem. Ecol. 2015, 3, 26. [Google Scholar] [CrossRef] [Green Version]
- Consortium, T.F. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 2003, 31, 172–175. [Google Scholar] [CrossRef]
- Kriventseva, E.V.; Kuznetsov, D.; Tegenfeldt, F.; Manni, M.; Dias, R.; Simão, F.A.; Zdobnov, E.M. OrthoDB v10: Sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2019, 47, D807–D811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A. FastTree 2–Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Elsik, C.G.; Worley, K.C.; Bennett, A.K.; Beye, M.; Camara, F.; Childers, C.P.; de Graaf, D.C.; Debyser, G.; Deng, J.; Devreese, B.; et al. Finding the missing honey bee genes: Lessons learned from a genome upgrade. BMC Genom. 2014, 15, 86. [Google Scholar] [CrossRef] [Green Version]
- Foret, S.; Maleszka, R. Function and evolution of a gene family encoding odorant binding-like proteins in a social insect, the honey bee (Apis mellifera). Genome Res. 2006, 16, 1404–1413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonasio, R.; Zhang, G.; Ye, C.; Mutti, N.S.; Fang, X.; Qin, N.; Donahue, G.; Yang, P.; Li, Q.; Li, C.; et al. Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science 2010, 329, 1068–1071. [Google Scholar] [CrossRef] [Green Version]
- McKenzie, S.K.; Oxley, P.R.; Kronauer, D.J.C. Comparative genomics and transcriptomics in ants provide new insights into the evolution and function of odorant binding and chemosensory proteins. BMC Genom. 2014, 15, 718. [Google Scholar] [CrossRef] [Green Version]
- Smith, C.D.; Zimin, A.; Holt, C.; Abouheif, E.; Benton, R.; Cash, E.; Croset, V.; Currie, C.R.; Elhaik, E.; Elsik, C.G.; et al. Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile). Proc. Natl. Acad. Sci. USA 2011, 108, 5673–5678. [Google Scholar] [CrossRef] [Green Version]
- Vieira, F.G.; Forêt, S.; He, X.; Rozas, J.; Field, L.; Zhou, J.-J. Unique Features of Odorant-Binding Proteins of the Parasitoid Wasp Nasonia vitripennis Revealed by Genome Annotation and Comparative Analyses. PLoS ONE 2012, 7, e43034. [Google Scholar] [CrossRef] [Green Version]
- Gong, D.-P.; Zhang, H.-J.; Zhao, P.; Xia, Q.-Y.; Xiang, Z.-H. The Odorant Binding Protein Gene Family from the Genome of Silkworm, Bombyx mori. BMC Genom. 2009, 10, 332. [Google Scholar] [CrossRef] [Green Version]
- Heliconius Genome Consortium. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 2012, 487, 94–98. [Google Scholar] [CrossRef] [Green Version]
- Zhan, S.; Reppert, S.M. MonarchBase: The monarch butterfly genome database. Nucleic Acids Res. 2012, 41, D758–D763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, R.; Lehane, S.; He, X.; Lehane, M.; Hertz-Fowler, C.; Berriman, M.; Pickett, J.A.; Field, L.M.; Zhou, J.-J. Characterisations of odorant-binding proteins in the tsetse fly Glossina morsitans morsitans. Cell. Mol. Life Sci. 2009, 67, 919–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawson, D.; Arensburger, P.; Atkinson, P.; Besansky, N.J.; Bruggner, R.V.; Butler, R.; Campbell, K.S.; Christophides, G.K.; Christley, S.; Dialynas, E.; et al. VectorBase: A data resource for invertebrate vector genomics. Nucleic Acids Res. 2009, 37, D583–D587. [Google Scholar] [CrossRef] [PubMed]
- Vieira, F.G.; Rozas, J. Comparative Genomics of the Odorant-Binding and Chemosensory Protein Gene Families across the Arthropoda: Origin and Evolutionary History of the Chemosensory System. Genome Biol. Evol. 2011, 3, 476–490. [Google Scholar] [CrossRef]
- Wang, J.; Hu, P.; Gao, P.; Tao, J.; Luo, Y. Antennal transcriptome analysis and expression profiles of olfactory genes in Anoplophora chinensis. Sci. Rep. 2017, 7, 15470. [Google Scholar] [CrossRef]
- Li, X.; Ju, Q.; Jie, W.; Li, F.; Jiang, X.; Hu, J.; Qu, M. Chemosensory Gene Families in Adult Antennae of Anomala corpulenta Motschulsky (Coleoptera: Scarabaeidae: Rutelinae). PLoS ONE 2015, 10, e0121504. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Q.; Zhao, H.; Ren, B. Identification and Comparison of Candidate Olfactory Genes in the Olfactory and Non-Olfactory Organs of Elm Pest Ambrostoma quadriimpressum (Coleoptera: Chrysomelidae) Based on Transcriptome Analysis. PLoS ONE 2016, 11, e0147144. [Google Scholar] [CrossRef] [Green Version]
- Bin, S.-Y.; Qu, M.-Q.; Li, K.-M.; Peng, Z.-Q.; Wu, Z.-Z.; Lin, J.-T. Antennal and abdominal transcriptomes reveal chemosensory gene families in the coconut hispine beetle, Brontispa longissima. Sci. Rep. 2017, 7, 2809. [Google Scholar] [CrossRef] [Green Version]
- Li, X.-M.; Zhu, X.-Y.; Wang, Z.-Q.; Wang, Y.; He, P.; Chen, G.; Sun, L.; Deng, D.-G.; Zhang, Y.-N. Candidate chemosensory genes identified in Colaphellus bowringi by antennal transcriptome analysis. BMC Genom. 2015, 16, 1028. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.-N.; Kang, K.; Xu, L.; Zhu, X.-Y.; Qian, J.-L.; Zhang, Z.-J.; He, P.; Li, X.-M. Deep sequencing of antennal transcriptome from Callosobruchus chinensis to characterize odorant binding protein and chemosensory protein genes. J. Stored Prod. Res. 2017, 74, 13–21. [Google Scholar] [CrossRef]
- Bin, S.-Y.; Qu, M.-Q.; Pu, X.-H.; Wu, Z.-Z.; Lin, J.-T. Antennal transcriptome and expression analyses of olfactory genes in the sweetpotato weevil Cylas formicarius. Sci. Rep. 2017, 7, 11073. [Google Scholar] [CrossRef] [Green Version]
- Andersson, M.N.; Grosse-Wilde, E.; Keeling, C.I.; Bengtsson, J.M.; Yuen, M.M.; Li, M.; Hillbur, Y.; Bohlmann, J.; Hansson, B.S.; Schlyter, F. Antennal transcriptome analysis of the chemosensory gene families in the tree killing bark beetles, Ips typographus and Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytinae). BMC Genom. 2013, 14, 198. [Google Scholar] [CrossRef] [Green Version]
- Keeling, C.I.; Yuen, M.M.; Liao, N.Y.; Docking, T.R.; Chan, S.K.; Taylor, G.A.; Palmquist, D.L.; Jackman, S.D.; Nguyen, A.; Li, M.; et al. Draft genome of the mountain pine beetle, Dendroctonus ponderosae Hopkins, a major forest pest. Genome Biol. 2013, 14, R27. [Google Scholar] [CrossRef] [Green Version]
- Yin, J.; Wang, C.; Fang, C.; Zhang, S.; Cao, Y.; Li, K.; Leal, W.S. Functional characterization of odorant-binding proteins from the scarab beetle Holotrichia oblita based on semiochemical-induced expression alteration and gene silencing. Insect Biochem. Mol. Biol. 2018, 104, 11–19. [Google Scholar] [CrossRef]
- Li, L.; Zhou, Y.-T.; Tan, Y.; Zhou, X.-R.; Pang, B.-P. Identification of odorant-binding protein genes in Galeruca daurica (Coleoptera: Chrysomelidae) and analysis of their expression profiles. Bull. Entomol. Res. 2017, 107, 550–561. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, L.; Cao, D.; Walker, W.B.; Zhang, Y.; Wang, G. Identification of candidate olfactory genes in Leptinotarsa decemlineata by antennal transcriptome analysis. Front. Ecol. Evol. 2015, 3, 60. [Google Scholar] [CrossRef] [Green Version]
- Yan, W.; Liu, L.; Qin, W.; Luo, Y.; Ma, X.; Haider, N.; Inayeh, M. Identification and tissue expression profiling of odorant binding protein genes in the red palm weevil, Rhynchophorus ferrugineus. SpringerPlus 2016, 5, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Robertson, H.M.; Baits, R.L.; Walden, K.K.; Wada-Katsumata, A.; Schal, C. Enormous expansion of the chemosensory gene repertoire in the omnivorous German cockroach Blattella germanica. J. Exp. Zool. Part B Mol. Dev. Evol. 2018, 330, 265–278. [Google Scholar] [CrossRef] [Green Version]
- Terrapon, N.; Li, C.; Robertson, H.M.; Ji, L.; Meng, X.; Booth, W.; Chen, Z.; Childers, C.; Glastad, K.; Gokhale, K.; et al. Molecular traces of alternative social organization in a termite genome. Nat. Commun. 2014, 5, 3636. [Google Scholar] [CrossRef] [Green Version]
- Petersen, T.N.; Søren Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; DiMaio, F.; Wang, R.Y.-R.; Kim, D.; Miles, C.; Brunette, T.; Thompson, J.; Baker, D. High-Resolution Comparative Modeling with RosettaCM. Structure 2013, 21, 1735–1742. [Google Scholar] [CrossRef] [Green Version]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Conway, P.; Tyka, M.D.; DiMaio, F.; Konerding, D.E.; Baker, D. Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci. Publ. Protein Soc. 2014, 23, 47–55. [Google Scholar] [CrossRef]
- Nivón, L.G.; Moretti, R.; Baker, D. A Pareto-Optimal Refinement Method for Protein Design Scaffolds. PLoS ONE 2013, 8, e59004. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Bradley, P.; Baker, D. Protein–Protein Docking with Backbone Flexibility. J. Mol. Biol. 2007, 373, 503–519. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.; Krissinel, E.B.; Leslie, A.G.W.; McCoy, A.; et al. Overview of theCCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Dippel, S.; Kollmann, M.; Oberhofer, G.; Montino, A.; Knoll, C.; Krala, M.; Rexer, K.-H.; Frank, S.; Kumpf, R.; Schachtner, J.; et al. Morphological and Transcriptomic Analysis of a Beetle Chemosensory System Reveals a Gnathal Olfactory Center. BMC Biol. 2016, 14, 90. [Google Scholar] [CrossRef] [Green Version]
- Roth, L.M.; Willis, E.R. Hygroreceptors in Coleoptera. J. Exp. Zool. 1951, 117, 451–487. [Google Scholar] [CrossRef]
- Zhang, S.-Q.; Che, L.-H.; Li, Y.; Liang, D.; Pang, H.; Ślipiński, A.; Zhang, P. Evolutionary history of Coleoptera revealed by extensive sampling of genes and species. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Andronopoulou, E.; Labropoulou, V.; Douris, V.; Woods, D.F.; Biessmann, H.; Iatrou, K. Specific interactions among odorant-binding proteins of the African malaria vector Anopheles gambiae. Insect Mol. Biol. 2006, 15, 797–811. [Google Scholar] [CrossRef]
- Wang, B.; Guan, L.; Zhong, T.; Li, K.; Yin, J.; Cao, Y. Potential Cooperations between Odorant-Binding Proteins of the Scarab Beetle Holotrichia oblita Faldermann (Coleoptera: Scarabaeidae). PLoS ONE 2013, 8, e84795. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T. UCSF Chimera?A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]




| Forward Primer | OBP | Reverse Primer |
|---|---|---|
| aaaccATGGCTGCGATGTCTGAGGC | OBP9A EcoRIrev | TTTGAATTCTCAGGGGAGAAAGTACTTTTCAGGATTG |
| aaaccATGGCGATGAGTGAAGCCC | OBP9B EcoRIrev | TTTGAATTCTTACGGTAAGAAGTATTTCTCGGGATTATCC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montino, A.; Balakrishnan, K.; Dippel, S.; Trebels, B.; Neumann, P.; Wimmer, E.A. Mutually Exclusive Expression of Closely Related Odorant-Binding Proteins 9A and 9B in the Antenna of the Red Flour Beetle Tribolium castaneum. Biomolecules 2021, 11, 1502. https://doi.org/10.3390/biom11101502
Montino A, Balakrishnan K, Dippel S, Trebels B, Neumann P, Wimmer EA. Mutually Exclusive Expression of Closely Related Odorant-Binding Proteins 9A and 9B in the Antenna of the Red Flour Beetle Tribolium castaneum. Biomolecules. 2021; 11(10):1502. https://doi.org/10.3390/biom11101502
Chicago/Turabian StyleMontino, Alice, Karthi Balakrishnan, Stefan Dippel, Björn Trebels, Piotr Neumann, and Ernst A. Wimmer. 2021. "Mutually Exclusive Expression of Closely Related Odorant-Binding Proteins 9A and 9B in the Antenna of the Red Flour Beetle Tribolium castaneum" Biomolecules 11, no. 10: 1502. https://doi.org/10.3390/biom11101502
APA StyleMontino, A., Balakrishnan, K., Dippel, S., Trebels, B., Neumann, P., & Wimmer, E. A. (2021). Mutually Exclusive Expression of Closely Related Odorant-Binding Proteins 9A and 9B in the Antenna of the Red Flour Beetle Tribolium castaneum. Biomolecules, 11(10), 1502. https://doi.org/10.3390/biom11101502