Single-Cell Transcriptome Analysis of H5N1-HA-Stimulated Alpaca PBMCs
Abstract
:1. Introduction
2. Materials and Methods
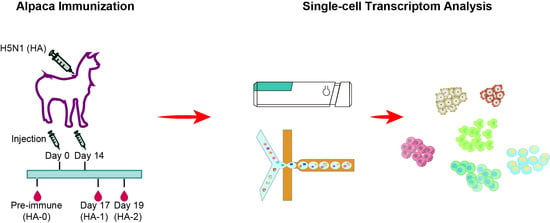
2.1. Alpaca Immunization
2.2. Serum Titer Assay
2.3. Peripheral Blood Mononuclear Cell (PBMC) Isolation
2.4. Single-Cell Library Construction and Sequencing
2.5. scRNA-Seq Data Processing
2.6. Data Integrating and Cell Clustering
2.7. Differentially Expressed Gene (DEG) Analysis
2.8. Gene Ontology Analysis
3. Results
3.1. Overview of the Transcriptome Landscape of Alpaca PBMCs
3.2. Monocyte Response to HA Immunization
3.3. B Cell Response to H5N1-HA Stimulation
3.4. The Transcriptome Landscape of T Cells in the Alpaca
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wille, M.; Barr, I.G. Resurgence of avian influenza virus. Science 2022, 376, 459–460. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Influenza at the Human-Animal Interface: Monthly Risk Assessment and Summary; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Skarlupka, A.L.; Bebin-Blackwell, A.-G.; Sumner, S.F.; Ross, T.M. Universal Influenza Virus Neuraminidase Vaccine Elicits Protective Immune Responses against Human Seasonal and Pre-pandemic Strains. J. Virol. 2021, 95, JVI0075921. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, E.; Neyts, J. Avian influenza A (H5N1) infection: Targets and strategies for chemotherapeutic intervention. Trends Pharmacol. Sci. 2007, 28, 280–285. [Google Scholar] [CrossRef]
- Berlanda Scorza, F.; Tsvetnitsky, V.; Donnelly, J.J. Universal influenza vaccines: Shifting to better vaccines. Vaccine 2016, 34, 2926–2933. [Google Scholar] [CrossRef] [Green Version]
- Subbarao, K.; Murphy, B.R.; Fauci, A.S. Development of effective vaccines against pandemic influenza. Immunity 2006, 24, 5–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nachbagauer, R.; Feser, J.; Naficy, A.; Bernstein, D.I.; Guptill, J.; Walter, E.B.; Berlanda-Scorza, F.; Stadlbauer, D.; Wilson, P.C.; Aydillo, T.; et al. A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat. Med. 2021, 27, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Clegg, C.H.; Rininger, J.A.; Baldwin, S.L. Clinical vaccine development for H5N1 influenza. Expert Rev. Vaccines 2013, 12, 767–777. [Google Scholar] [CrossRef]
- Hussen, J.; Schuberth, H.J. Recent Advances in Camel Immunology. Front. Immunol. 2021, 11, 614150. [Google Scholar] [CrossRef]
- Kulkarni, S.S.; Falzarano, D. Unique aspects of adaptive immunity in camelids and their applications. Mol. Immunol. 2021, 134, 102–108. [Google Scholar] [CrossRef]
- Blanc, M.R.; Anouassi, A.; Abed, M.A.; Tsikis, G.; Canepa, S.; Labas, V.; Belghazi, M.; Bruneau, G. A one-step exclusion-binding procedure for the purification of functional heavy-chain and mammalian-type γ-globulins from camelid sera. Biotechnol. Appl. Biochem. 2009, 54, 207–212. [Google Scholar] [CrossRef]
- Hamers-Casterman, C.; Atarhouch, T.; Muyldermans, S.; Robinson, G.; Hammers, C.; Songa, E.B.; Bendahman, N.; Hammers, R. Naturally occurring antibodies devoid of light chains. Nature 1993, 363, 446–448. [Google Scholar] [CrossRef] [PubMed]
- Del Rosario, J.M.M.; Smith, M.; Zaki, K.; Risley, P.; Temperton, N.; Engelhardt, O.G.; Collins, M.; Takeuchi, Y.; Hufton, S.E. Protection From Influenza by Intramuscular Gene Vector Delivery of a Broadly Neutralizing Nanobody Does Not Depend on Antibody Dependent Cellular Cytotoxicity. Front. Immunol. 2020, 11, 627. [Google Scholar] [CrossRef] [PubMed]
- Vanlandschoot, P.; Stortelers, C.; Beirnaert, E.; Ibañez, L.I.; Schepens, B.; Depla, E.; Saelens, X. Nanobodies®: New ammunition to battle viruses. Antivir. Res. 2011, 92, 389–407. [Google Scholar] [CrossRef] [PubMed]
- Muyldermans, S. Nanobodies: Natural single-domain antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef] [Green Version]
- Topliff, C.L.; Alkheraif, A.A.; Kuszynski, C.A.; Davis, W.C.; Steffen, D.J.; Schmitz, J.A.; Eskridge, K.M.; Charleston, B.; Henningson, J.N.; Kelling, C.L. Experimental acute infection of alpacas with Bovine viral diarrhea virus 1 subgenotype b alters peripheral blood and GALT leukocyte subsets. J. Vet. Diagn. Investig. 2017, 29, 186–192. [Google Scholar] [CrossRef] [Green Version]
- Zidan, M.; Schuberth, H.J.; Pabst, R. Immunohistology of the splenic compartments of the one humped camel (Camelus dromedarius). Vet. Immunol. Immunopathol. 2000, 74, 17–29. [Google Scholar] [CrossRef]
- Ungar-Waron, H.; Yagil, R.; Brenner, J.; Paz, R.; Partosh, N.; van Creveld, C.; Lubashevsky, E.; Trainin, Z. Reactions of peripheral blood mononuclear cells (PBMC) of camels with monoclonal antibodies against ruminant leukocytes. Comp. Immunol. Microbiol. Infect. Dis. 2003, 26, 137–143. [Google Scholar] [CrossRef]
- Henry, K.A.; van Faassen, H.; Harcus, D.; Marcil, A.; Hill, J.J.; Muyldermans, S.; Mackenzie, C.R. Llama peripheral B-cell populations producing conventional and heavy chain-only IgG subtypes are phenotypically indistinguishable but immunogenetically distinct. Immunogenetics 2019, 71, 307–320. [Google Scholar] [CrossRef]
- Papalexi, E.; Satija, R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 2018, 18, 35–45. [Google Scholar] [CrossRef]
- Olsen, T.K.; Baryawno, N. Introduction to Single-Cell RNA Sequencing. Curr. Protoc. Mol. Biol. 2018, 122, e57. [Google Scholar] [CrossRef]
- Satija, R.; Farrell, J.A.; Gennert, D.; Schier, A.F.; Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015, 33, 495–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M.; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef] [PubMed]
- Szabo, P.A.; Levitin, H.M.; Miron, M.; Snyder, M.E.; Senda, T.; Yuan, J.; Cheng, Y.L.; Bush, E.C.; Dogra, P.; Thapa, P.; et al. Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat. Commun. 2019, 10, 4706. [Google Scholar] [CrossRef] [Green Version]
- Among, T.; Clusters, G.; Yu, G. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Lamichhane, P.P.; Samarasinghe, A.E. The Role of Innate Leukocytes during Influenza Virus Infection. J. Immunol. Res. 2019, 2019, 8028725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hedges, J.F.; Kimmel, E.; Snyder, D.T.; Jerome, M.; Jutila, M.A. Solute Carrier 11A1 Is Expressed by Innate Lymphocytes and Augments Their Activation. J. Immunol. References 2022, 190, 4263–4273. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Shively, J.E. Acceleration of Bone Repair in NOD/SCID Mice by Human Monoosteophils, Novel LL-37-Activated Monocytes. PLoS ONE 2013, 8, e67649. [Google Scholar] [CrossRef] [Green Version]
- Vandestienne, M.; Zhang, Y.; Santos-Zas, I.; Al-Rifai, R.; Joffre, J.; Giraud, A.; Laurans, L.; Esposito, B.; Pinet, F.; Bruneval, P.; et al. TREM-1 orchestrates angiotensin II-induced monocyte trafficking and promotes experimental abdominal aortic aneurysm. J. Clin. Investig. 2021, 131, e142468. [Google Scholar] [CrossRef]
- Ando, K.; Hasegawa, K.; Shindo, K.-I.; Furusawa, T.; Fujino, T.; Kikugawa, K.; Nakano, H.; Takeuchi, O.; Akira, S.; Akiyama, T.; et al. Human lactoferrin activates NF-jB through the Toll-like receptor 4 pathway while it interferes with the lipopolysaccharide-stimulated TLR4 signaling. FEBS J. 2010, 277, 2051–2066. [Google Scholar] [CrossRef]
- Ghavami, S.; Chitayat, S.; Hashemi, M.; Eshraghi, M.; Chazin, W.J.; Halayko, A.J.; Kerkhoff, C. S100A8/A9: A Janus-faced molecule in cancer therapy and tumorgenesis. Eur. J. Pharmacol. 2009, 625, 73–83. [Google Scholar] [CrossRef]
- Manolakis, A.C.; Kapsoritakis, A.N.; Tiaka, E.K.; Potamianos, S.P. Calprotectin, Calgranulin, C, and Other Members of the S100 Protein Family in Inflammatory Bowel Disease. Dig. Dis. Sci. 2011, 56, 1601–1611. [Google Scholar] [CrossRef] [PubMed]
- Foell, D.; Wittkowski, H.; Kessel, C.; Lüken, A.; Weinhage, T.; Varga, G.; Vogl, T.; Wirth, T.; Viemann, D.; Björk, P.; et al. Proinflammatory S100A12 can activate human monocytes via toll-like receptor 4. Am. J. Respir. Crit. Care Med. 2013, 187, 1324–1334. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Wang, Y.; Zhao, S.; Zhang, Y.; Chen, Y.; Zhao, X.; Liu, L.; Sun, S.; Zhang, L.; Ye, B.; et al. MICAL1 facilitates breast cancer cell proliferation via ROS-sensitive ERK/cyclin D pathway. J. Cell. Mol. Med. 2018, 22, 3108–3118. [Google Scholar] [CrossRef]
- Xu, J.; Chi, F.; Guo, T.; Punj, V.; Paul Lee, W.N.; French, S.W.; Tsukamoto, H. NOTCH reprograms mitochondrial metabolism for proinflammatory macrophage activation. J. Clin. Investig. 2015, 125, 1579–1590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singla, D.K.; Wang, J.; Singla, R. Primary human monocytes differentiate into M2 macrophages and involve notch-1 pathway. Can. J. Physiol. Pharmacol. 2016, 95, 288–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gamrekelashvili, J.; Kapanadze, T.; Sablotny, S.; Ratiu, C.; Dastagir, K.; Lochner, M.; Karbach, S.; Wenzel, P.; Sitnow, A.; Fleig, S.; et al. Notch and TLR signaling coordinate monocyte cell fate and inflammation. Elife 2020, 9, e57007. [Google Scholar] [CrossRef]
- Kaiser, A.; Schmidt, M.; Huber, O.; Frietsch, J.J.; Scholl, S.; Heidel, F.H.; Hochhaus, A.; Müller, J.P.; Ernst, T. SIRT7: An influence factor in healthy aging and the development of age-dependent myeloid stem-cell disorders. Leukemia 2020, 34, 2206–2216. [Google Scholar] [CrossRef]
- Li, H.; Zhao, X.-K.; Cheng, Y.-J.; Zhang, Q.; Wu, J.; Lu, S.; Zhang, W.; Liu, Y.; Zhou, M.-Y.; Wang, Y.; et al. Gasdermin D-mediated hepatocyte pyroptosis expands inflammatory responses that aggravate acute liver failure by upregulating monocyte chemotactic protein 1/CC chemokine receptor-2 to recruit macrophages. World J. Gastroenterol. 2019, 25, 6527–6540. [Google Scholar] [CrossRef]
- Zhou, B.; Liu, J.; Zeng, L.; Zhu, S.; Wang, H.; Billiar, T.R.; Kroemer, G.; Klionsky, D.J.; Zeh, H.J.; Jiang, J.; et al. Extracellular SQSTM1 mediates bacterial septic death in mice through insulin receptor signalling. Nat. Microbiol. 2020, 5, 1576–1587. [Google Scholar] [CrossRef]
- Kapellos, T.S.; Bonaguro, L.; Gemünd, I.; Reusch, N.; Saglam, A.; Hinkley, E.R.; Schultze, J.L. Human monocyte subsets and phenotypes in major chronic inflammatory diseases. Front. Immunol. 2019, 10, 2035. [Google Scholar] [CrossRef]
- Krammer, F. The human antibody response to influenza A virus infection and vaccination. Nat. Rev. Immunol. 2019, 19, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.W.; Thuy, P.X.; Lee, J.W.; Moon, E.Y. CXCR4 promotes B cell viability by the cooperation of nuclear factor (erythroid-derived 2)-like 2 and hypoxia-inducible factor-1α under hypoxic conditions. Cell Death Dis. 2021, 12, 330. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Waite, J.; Brewer, F.; Sunshine, M.J.; Littman, D.R.; Zou, Y.R. The role of CXCR4 in maintaining peripheral B cell compartments and humoral immunity. J. Exp. Med. 2004, 200, 1145–1156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, H.; Morand, F.E. The Role of GILZ in Anti-Inflammatory and Immunosuppressive Actions of Glucocorticoids. In Glucocorticoids—New Recognition of Our Familiar Friend; InTech: Rijeka, Croatia, 2012; pp. 175–192. [Google Scholar] [CrossRef]
- Morgan, D.; Tergaonkar, V. Unraveling B cell trajectories at single cell resolution. Trends Immunol. 2022, 43, 210–229. [Google Scholar] [CrossRef] [PubMed]
- Huber, C.; Mårtensson, A.; Bokoch, G.M.; Nemazee, D.; Gavin, A.L. FGD2, a CDC42-specific exchange factor expressed by antigen-presenting cells, localizes to early endosomes and active membrane ruffles. J. Biol. Chem. 2008, 283, 34002–34012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Junco, J.J.; Zorman, B.; Gant, V.U.; Muñoz, J.; Lacorazza, H.D.; Sumazin, P.; Rabin, K.R. CRLF2 overexpression results in reduced B-cell differentiation and upregulated E2F signaling in the Dp16 mouse model of Down syndrome. Exp. Hematol. 2022, 110, 34–38. [Google Scholar] [CrossRef]
- Schmidlin, H.; Diehl, S.A.; Nagasawa, M.; Scheeren, F.A.; Schotte, R.; Uittenbogaart, C.H.; Spits, H.; Blom, B. Spi-B inhibits human plasma cell differentiation by repressing BLIMP1 and XBP-1 expression. Blood 2008, 112, 1804–1812. [Google Scholar] [CrossRef] [Green Version]
- Jash, A.; Wang, Y.; Weisel, F.J.; Scharer, C.D.; Boss, J.M.; Shlomchik, M.J.; Bhattacharya, D. ZBTB32 Restricts the Duration of Memory B Cell Recall Responses. J. Immunol. 2016, 197, 1159–1168. [Google Scholar] [CrossRef] [Green Version]
- Tsui, C.; Maldonado, P.; Montaner, B.; Borroto, A.; Alarcon, B.; Bruckbauer, A.; Martinez-Martin, N.; Batista, F.D. Dynamic reorganisation of intermediate filaments coordinates early B-cell activation. Life Sci. Alliance 2018, 1, e201800060. [Google Scholar] [CrossRef]
- Wang, X.H.; Du, H.; Li, L.; Shao, D.F.; Zhong, X.Y.; Hu, Y.; Liu, Y.Q.; Xing, X.F.; Cheng, X.J.; Guo, T.; et al. Increased expression of S100A6 promotes cell proliferation in gastric cancer cells. Oncol. Lett. 2017, 13, 222–230. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, F.; Umeda, Y.; Shimamoto, S.; Tsuchiya, M.; Tokumitsu, H.; Tokuda, M.; Kobayashi, R. S100 proteins modulate protein phosphatase 5 function: A link between Ca2+ signal transduction and protein dephosphorylation. J. Biol. Chem. 2012, 287, 13787–13798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charlestin, V.; Fulkerson, D.; Arias Matus, C.E.; Walker, Z.T.; Carthy, K.; Littlepage, L.E. Aquaporins: New players in breast cancer progression and treatment response. Front. Oncol. 2022, 12, 988119. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Sanchez, H.N.; Zan, H.; Casali, P. Genome-wide analysis reveals selective modulation of microRNAs and mRNAs by histone deacetylase inhibitor in B cells induced to undergo class-switch DNA recombination and plasma cell differentiation. Front. Immunol. 2015, 6, 627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kassambara, A.; Herviou, L.; Ovejero, S.; Jourdan, M.; Thibaut, C.; Vikova, V.; Pasero, P.; Elemento, O.; Moreaux, J. RNA-sequencing data-driven dissection of human plasma cell differentiation reveals new potential transcription regulators. Leukemia 2021, 35, 1451–1462. [Google Scholar] [CrossRef] [PubMed]
- Castro, C.D.; Flajnik, M.F. Putting J Chain Back on the Map: How Might Its Expression Define Plasma Cell Development? J. Immunol. 2014, 193, 3248–3255. [Google Scholar] [CrossRef] [Green Version]
- Erlandsson, L.; Akerblad, P.; Vingsbo-Lundberg, C.; Kallberg, E.; Lycke, N.; Leanderson, T. Joining Chain-expressing and-nonexpressing B Cell Populations in the Mouse. J. Exp. Med. 2001, 194, 557–570. [Google Scholar] [CrossRef]
- Xu, A.Q.; Barbosa, R.R.; Calado, D.P. Genetic timestamping of plasma cells in vivo reveals tissue-specific homeostatic population turnover. Elife 2020, 9, e59850. [Google Scholar] [CrossRef]
- Andreani, V.; Ramamoorthy, S.; Pandey, A.; Lupar, E.; Nutt, S.L.; Lämmermann, T.; Grosschedl, R. Cochaperone Mzb1 is a key effector of Blimp1 in plasma cell differentiation and β1-integrin function. Proc. Natl. Acad. Sci. USA 2018, 115, E9630–E9639. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Li, Z. Endoplasmic reticulum HSP90bl (gp96, grp94) optimizes B-cell function via chaperoning integrin and TLR but not immunoglobulin. Blood 2008, 112, 1223–1230. [Google Scholar] [CrossRef]
- Koutsakos, M.; Kedzierska, K.; Subbarao, K. Immune Responses to Avian Influenza Viruses. J. Immunol. 2019, 202, 382–391. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Loh, L.; Kedzierski, L.; Kedzierska, K. Avian influenza viruses, inflammation, and CD8+ T cell immunity. Front. Immunol. 2016, 7, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Comor, L.; Dolinska, S.; Bhide, K.; Pulzova, L.; Munguía, I.J.; Bencurova, E.; Flachbartova, Z.; Potocnakova, L.; Kanova, E.; Bhide, M. Joining the in vitro immunization of alpaca lymphocytes and phage display: Rapid and cost effective pipeline for sdAb synthesis. Microb. Cell Fact. 2017, 16, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chow, K.M.; Whiteheart, S.W.; Smiley, J.R.; Sharma, S.; Boaz, K.; Coleman, M.J.; Maynard, A.; Hersh, L.B.; Kooi, C.W.V. Immunization of Alpacas (Lama pacos) with Protein Antigens and Production of Antigen-specific Single Domain Antibodies. J. Vis. Exp. 2019, 143, e58471. [Google Scholar] [CrossRef] [Green Version]
- Pulendran, B.; Ahmed, R. Translating innate immunity into immunological memory: Implications for vaccine development. Cell 2006, 124, 849–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biondo, C.; Lentini, G.; Beninati, C.; Teti, G. The dual role of innate immunity during influenza. Biomed. J. 2019, 42, 8–18. [Google Scholar] [CrossRef]
- Hoeve, M.A.; Nash, A.A.; Jackson, D.; Randall, R.E.; Dransfield, I. Influenza virus A infection of human monocyte and macrophage subpopulations reveals increased susceptibility associated with cell differentiation. PLoS ONE 2012, 7, e29443. [Google Scholar] [CrossRef] [Green Version]
- Tisoncik, J.R.; Korth, M.J.; Simmons, C.P.; Farrar, J.; Martin, T.R.; Katze, M.G. Into the Eye of the Cytokine Storm. Microbiol. Mol. Biol. Rev. 2012, 76, 16–32. [Google Scholar] [CrossRef] [Green Version]
- Van Furth, R. Monocyte production during inflammation. Comp. Immunol. Microbiol. Infect. Dis. 1985, 8, 205–211. [Google Scholar] [CrossRef]
- Van Den Broek, T.; Borghans, J.A.M.; Van Wijk, F. The full spectrum of human naive T cells. Nat. Rev. Immunol. 2018, 18, 363–373. [Google Scholar] [CrossRef]
- Caccamo, N.; Joosten, S.A.; Ottenhoff, T.H.M.; Dieli, F. Atypical Human Effector/Memory CD4+ T Cells with a Naive-Like Phenotype. Front. Immunol. 2018, 9, 2832. [Google Scholar] [CrossRef] [Green Version]
- Tena-Coki, N.G.; Scriba, T.J.; Peteni, N.; Eley, B.; Wilkinson, R.J.; Andersen, P.; Hanekom, W.A.; Kampmann, B. CD4 and CD8 T-cell responses to mycobacterial antigens in African children. Am. J. Respir. Crit. Care Med. 2010, 182, 120–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soares, A.P.; Kwong Chung, C.K.C.; Choice, T.; Hughes, E.J.; Jacobs, G.; Van Rensburg, E.J.; Khomba, G.; De Kock, M.; Lerumo, L.; Makhethe, L.; et al. Longitudinal changes in CD4+ T-cell memory responses induced by BCG vaccination of newborns. J. Infect. Dis. 2013, 207, 1084–1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soares, A.P.; Scriba, T.J.; Joseph, S.; Harbacheuski, R.; Murray, R.A.; Gelderbloem, S.J.; Hawkridge, A.; Hussey, G.D.; Maecker, H.; Kaplan, G.; et al. Bacillus Calmette-Guérin Vaccination of Human Newborns Induces T Cells with Complex Cytokine and Phenotypic Profiles. J. Immunol. 2008, 180, 3569–3577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Dement-Brown, J.; Liao, P.J.; Mazo, I.; Mills, F.; Kraus, Z.; Fitzsimmons, S.; Tolnay, M. Fc receptor-like 4 and 5 define human atypical memory B cells. Int. Immunol. 2020, 32, 755–770. [Google Scholar] [CrossRef] [PubMed]
- Sutton, H.J.; Aye, R.; Idris, A.H.; Vistein, R.; Nduati, E.; Kai, O.; Mwacharo, J.; Li, X.; Gao, X.; Andrews, T.D.; et al. Atypical B cells are part of an alternative lineage of B cells that participates in responses to vaccination and infection in humans. Cell Rep. 2021, 34, 108684. [Google Scholar] [CrossRef] [PubMed]
- Mazutis, L.; Gilbert, J.; Ung, W.; A Weitz, D.; Griffiths, A.D.; Heyman, J.A. Single-cell analysis and sorting using droplet-based microfluidics. Nat. Protoc. 2014, 8, 870–891. [Google Scholar] [CrossRef] [PubMed]
- Gérard, A.; Woolfe, A.; Mottet, G.; Reichen, M.; Castrillon, C.; Menrath, V.; Ellouze, S.; Poitou, A.; Doineau, R.; Briseno-Roa, L.; et al. High-throughput single-cell activity-based screening and sequencing of antibodies using droplet microfluidics. Nat. Biotechnol. 2020, 38, 715–721. [Google Scholar] [CrossRef]
- Lyu, M.; Shi, X.; Liu, X.; Liu, Y.; Zhu, X.; Liao, L.; Zhao, H.; Sun, N.; Wang, S.; Chen, L.; et al. Generation and Screening of Antigen-Specific Nanobodies from Mammalian Cells Expressing the BCR Repertoire Library Using Droplet-Based Microfluidics. Anal. Chem. 2022, 94, 7970–7980. [Google Scholar] [CrossRef]
- Guo, X.; Chen, F.; Gao, F.; Li, L.; Liu, K.; You, L.; Hua, C.; Yang, F.; Liu, W.; Peng, C.; et al. CNSA: A data repository for archiving omics data. Database 2020, 2020, baaa055. [Google Scholar] [CrossRef]
- Chen, F.; You, L.; Yang, F.; Wang, L.; Guo, X.; Gao, F.; Hua, C. CNGBdb: China National GeneBank DataBase. Heredity 2020, 42, 799–809. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lyu, M.; Shi, X.; Liu, Y.; Zhao, H.; Yuan, Y.; Xie, R.; Gu, Y.; Dong, Y.; Wang, M. Single-Cell Transcriptome Analysis of H5N1-HA-Stimulated Alpaca PBMCs. Biomolecules 2023, 13, 60. https://doi.org/10.3390/biom13010060
Lyu M, Shi X, Liu Y, Zhao H, Yuan Y, Xie R, Gu Y, Dong Y, Wang M. Single-Cell Transcriptome Analysis of H5N1-HA-Stimulated Alpaca PBMCs. Biomolecules. 2023; 13(1):60. https://doi.org/10.3390/biom13010060
Chicago/Turabian StyleLyu, Menghua, Xuyang Shi, Yang Liu, Hongyan Zhao, Yue Yuan, Run Xie, Ying Gu, Yuliang Dong, and Meiniang Wang. 2023. "Single-Cell Transcriptome Analysis of H5N1-HA-Stimulated Alpaca PBMCs" Biomolecules 13, no. 1: 60. https://doi.org/10.3390/biom13010060
APA StyleLyu, M., Shi, X., Liu, Y., Zhao, H., Yuan, Y., Xie, R., Gu, Y., Dong, Y., & Wang, M. (2023). Single-Cell Transcriptome Analysis of H5N1-HA-Stimulated Alpaca PBMCs. Biomolecules, 13(1), 60. https://doi.org/10.3390/biom13010060