Identification of Transcription Factors and the Regulatory Genes Involved in Triacylglycerol Accumulation in the Unicellular Red Alga Cyanidioschyzon merolae
Abstract
:1. Introduction
2. Results
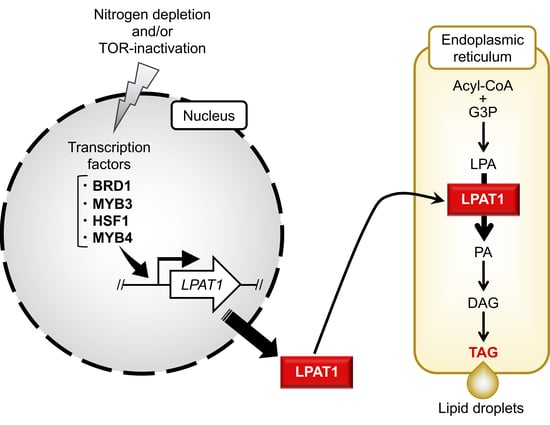
2.1. Identification of Candidate TFs Involved in TAG Accumulation
2.2. Effect of TF Overexpression on Lipid Droplet Formation and TAG Accumulation
2.3. Comparison of TAG Synthesis-Related Gene Expression in BRD1ox, MYB3ox, HSF1ox, and MYB4ox
2.4. TAG Accumulation in LPAT1 Overexpression Strain
3. Discussion
4. Materials and Methods
4.1. Strain and Growth Conditions
4.2. Construction of Overexpression Strains
4.3. Immunoblot Analysis
4.4. Lipid Analysis
4.5. RNA Preparation
4.6. Quantitative Real-Time PCR Analysis
4.7. RNA-Seq Analysis
4.8. Indirect Immuno-Fluorescence Microscopy Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chisti, Y. Biodiesel from microalgae. Biotechnol. Adv. 2007, 25, 294–306. [Google Scholar] [CrossRef] [PubMed]
- Chisti, Y. Biodiesel from microalgae beats bioethanol. Trends Biotechnol. 2008, 26, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Parmar, A.; Singh, N.K.; Pandey, A.; Gnansounou, E.; Madamwar, D. Cyanobacteria and microalgae: A positive prospect for biofuels. Bioresour. Technol. 2011, 102, 10163–10172. [Google Scholar] [CrossRef]
- Matsuzaki, M.; Misumi, O.; Shin-I, T.; Maruyama, S.; Takahara, M.; Miyagishima, S.Y.; Mori, T.; Nishida, K.; Yagisawa, F.; Nishida, K.; et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 2014, 428, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Imamura, S.; Kawase, Y.; Kobayashi, I.; Sone, T.; Era, A.; Miyagishima, S.Y.; Shimojima, M.; Ohta, H.; Tanaka, K. Target of rapamycin (TOR) plays a critical role in triacylglycerol accumulation in microalgae. Plant Mol. Biol. 2015, 89, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Sabatini, D.M. mTOR signaling in growth, metabolism, and disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [Green Version]
- Heitman, J.; Movva, N.R.; Hall, M.N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 1991, 253, 905–909. [Google Scholar] [CrossRef]
- Imamura, S.; Ishiwata, A.; Watanabe, S.; Yoshikawa, H.; Tanaka, K. Expression of budding yeast FKBP12 confers rapamycin susceptibility to the unicellular red alga Cyanidioschyzon merolae. Biochem. Biophys. Res. Commun. 2013, 439, 264–269. [Google Scholar] [CrossRef]
- Imamura, S.; Taki, K.; Tanaka, K. Construction of a rapamycin-susceptible strain of the unicellular red alga Cyanidioschyzon merolae for analysis of the target of rapamycin (TOR) function. J. Gen. Appl. Microbiol. 2017, 63, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Imamura, S.; Kawase, Y.; Kobayashi, I.; Shimojima, M.; Ohta, H.; Tanaka, K. TOR (target of rapamycin) is a key regulator of triacylglycerol accumulation in microalgae. Plant Signal. Behav. 2016, 11, e1149285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukaida, S.; Ogawa, T.; Ohishi, K.; Tanizawa, Y.; Ohta, D.; Arita, M. The effect of rapamycin on biodiesel-producing protist Euglena gracilis. Biosci. Biotechnol. Biochem. 2016, 80, 1223–1229. [Google Scholar] [CrossRef] [Green Version]
- Prioretti, L.; Carriere, F.; Field, B.; Avilan, L.; Montané, M.H.; Menand, B.; Gontero, B. Targeting TOR signaling for enhanced lipid productivity in algae. Biochimie 2019, 169, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Pancha, I.; Chokshi, K.; Tanaka, K.; Imamura, S. Microalgal Target of Rapamycin (TOR): A Central Regulatory Hub for Growth, Stress Response and Biomass Production. Plant Cell Physiol. 2020, 61, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.Y.; Benning, C. Triacylglycerol Accumulation in Photosynthetic Cells in Plants and Algae. Subcell. Biochem. 2016, 86, 179–205. [Google Scholar]
- Fukuda, S.; Hirasawa, E.; Takemura, T.; Takahashi, S.; Chokshi, K.; Pancha, I.; Tanaka, K.; Imamura, S. Accelerated triacylglycerol production without growth inhibition by overexpression of a glycerol-3-phosphate acyltransferase in the unicellular red alga Cyanidioschyzon merolae. Sci. Rep. 2018, 8, 12410. [Google Scholar] [CrossRef] [Green Version]
- de Nadal, E.; Ammerer, G.; Posas, F. Controlling gene expression in response to stress. Nat. Rev. Genet. 2011, 12, 833–845. [Google Scholar] [CrossRef] [PubMed]
- Boyle, N.R.; Page, M.D.; Liu, B.; Blaby, I.K.; Casero, D.; Kropat, J.; Cokus, S.J.; Hong-Hermesdorf, A.; Shaw, J.; Karpowicz, S.J.; et al. Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J. Biol. Chem. 2012, 287, 15811–15825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goncalves, E.C.; Koh, J.; Zhu, N.; Yoo, M.J.; Chen, S.; Matsuo, T.; Johnson, J.V.; Rathinasabapathi, B. Nitrogen starvation-induced accumulation of triacylglycerol in the green algae: Evidence for a role for ROC40, a transcription factor involved in circadian rhythm. Plant J. 2016, 85, 743–757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pancha, I.; Shima, H.; Higashitani, N.; Igarashi, K.; Higashitani, A.; Tanaka, K.; Imamura, S. Target of rapamycin-signaling modulates starch accumulation via glycogenin phosphorylation status in the unicellular red alga Cyanidioschyzon merolae. Plant J. 2019, 97, 485–499. [Google Scholar] [CrossRef] [Green Version]
- Fujii, G.; Imamura, S.; Hanaoka, M.; Tanaka, K. Nuclear-encoded chloroplast RNA polymerase sigma factor SIG2 activates chloroplast-encoded phycobilisome genes in a red alga, Cyanidioschyzon merolae. FEBS Lett. 2013, 587, 3354–3359. [Google Scholar] [CrossRef] [Green Version]
- Taki, K.; Sone, T.; Kobayashi, Y.; Watanabe, S.; Imamura, S.; Tanaka, K. Construction of a URA5.3 deletion strain of the unicellular red alga Cyanidioschyzon merolae: A backgroundless host strain for transformation experiments. J. Gen. Appl. Microbiol. 2015, 61, 211–214. [Google Scholar] [CrossRef] [Green Version]
- Yagisawa, F.; Nishida, K.; Yoshida, M.; Ohnuma, M.; Shimada, T.; Fujiwara, T.; Yoshida, Y.; Misumi, O.; Kuroiwa, H.; Kuroiwa, T. Identification of novel proteins in isolated polyphosphate vacuoles in the primitive red alga Cyanidioschyzon merolae. Plant J. 2009, 60, 882–893. [Google Scholar] [CrossRef]
- Musselman, C.A.; Lalonde, M.E.; Côté, J.; Kutateladze, T.G. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 2012, 19, 1218–1227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz, C.F.; Weusthuis, R.A.; D’Adamo, S.; Wijffels, R.H. Effect of Single and Combined Expression of Lysophosphatidic Acid Acyltransferase, Glycerol-3-Phosphate Acyltransferase, and Diacylglycerol Acyltransferase on Lipid Accumulation and Composition in Neochloris oleoabundans. Front. Plant Sci. 2019, 10, 1573. [Google Scholar] [CrossRef] [PubMed]
- Iwai, M.; Ikeda, K.; Shimojima, M.; Ohta, H. Enhancement of extraplastidic oil synthesis in Chlamydomonas reinhardtii using a type-2 diacylglycerol acyltransferase with a phosphorus starvation-inducible promoter. Plant Biotechnol. J. 2014, 12, 808–819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.W.; Cen, S.Y.; Liu, Y.H.; Balamurugan, S.; Zheng, X.Y.; Alimujiang, A.; Yang, W.D.; Liu, J.S.; Li, H.Y. A type 2 diacylglycerol acyltransferase accelerates the triacylglycerol biosynthesis in heterokont oleaginous microalga Nannochloropsis oceanica. J. Biotechnol. 2016, 229, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Klaitong, P.; Fa-Aroonsawat, S.; Chungjatupornchai, W. Accelerated triacylglycerol production and altered fatty acid composition in oleaginous microalga Neochloris oleoabundans by overexpression of diacylglycerol acyltransferase 2. Microb. Cell Fact. 2017, 16, 61. [Google Scholar] [CrossRef]
- Imamura, S.; Terashita, M.; Ohnuma, M.; Maruyama, S.; Minoda, A.; Weber, A.P.; Inouye, T.; Sekine, Y.; Fujita, Y.; Omata, T.; et al. Nitrate assimilatory genes and their transcriptional regulation in a unicellular red alga Cyanidioschyzon merolae: Genetic evidence for nitrite reduction by a sulfite reductase-like enzyme. Plant Cell Physiol. 2010, 51, 707–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takemura, T.; Kobayashi, Y.; Imamura, S.; Tanaka, K. Top starch plating method for the efficient cultivation of unicellular red alga Cyanidioschyzon merolae. Bioprotocol 2019, 9, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imamura, S.; Yoshihara, S.; Nakano, S.; Shiozaki, N.; Yamada, A.; Tanaka, K.; Takahashi, H.; Asayama, M.; Shirai, M. Purification, characterization, and gene expression of all sigma factors of RNA polymerase in a cyanobacterium. J. Mol. Biol. 2003, 325, 857–872. [Google Scholar] [CrossRef]
- Imamura, S.; Hanaoka, M.; Tanaka, K. The plant-specific TFIIB-related protein, pBrp, is a general transcription factor for RNA polymerase I. EMBO J. 2008, 27, 2317–2327. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Gene Number ¹ | Predicted Enzyme | Ratio (vs. TFc) | |||
|---|---|---|---|---|---|
| BRD1ox (CMK212Cox) | MYB3ox (CML101Cox) | HSF 1ox (CML277Cox) | MYB4ox (CMO347Cox) | ||
| CMS299C | Biotin carboxylase, chloroplast precursor | 0.9 | 1.2 | 1.5 | 1.2 |
| CMT420C | Malonyl-CoA ACP transacylase | 1.2 | 1.5 | 1.4 | 2.0 |
| CMM286C | 3-ketoacyl-ACP synthase | 1.9 | 2.2 | 2.4 | 1.9 |
| CML329C | 3-ketoacyl-ACP synthase | 1.0 | 1.0 | 1.0 | 1.0 |
| CMD118C | 3-ketoacyl-ACP synthase | 0.9 | 1.4 | 1.1 | 1.5 |
| CMS393C | 3-keotacyl-ACP reductase | 1.0 | 1.4 | 1.5 | 1.7 |
| CMI240C | 3-hydroxyacyll-ACP dehydratase | 0.6 | 1.0 | 0.8 | 1.2 |
| CMT381C | Enoyl-ACP reductase | 1.3 | 1.5 | 1.6 | 1.6 |
| CMJ027C | Glycerol-3-phosphate acyltransferase | 1.1 | 1.5 | 1.9 | 2.1 |
| CMA017C | Glycerol-3-phosphate acyltransferase | 1.0 | 1.4 | 1.4 | 1.8 |
| CMK217C | Glycerol-3-phosphate acyltransferase | 1.1 | 1.2 | 1.3 | 2.1 |
| CME109C | Lysophosphatidic acid acyltransferase | 1.0 | 1.3 | 1.4 | 1.9 |
| CMF185C | Lysophosphatidic acid acyltransferase | 0.9 | 1.0 | 1.2 | 1.8 |
| CMJ021C | Lysophosphatidic acid acyltransferase (LPAT1) | 39.9 | 2.0 | 133.5 | 13.0 |
| CMR054C | Phosphatidic acid phosphatase | 0.8 | 1.2 | 1.7 | 1.6 |
| CMR488C | Phosphatidic acid phosphatase | 1.5 | 1.6 | 2.3 | 1.8 |
| CMQ199C | Diacylglycerol acyltransferase | 1.0 | 1.2 | 1.3 | 1.9 |
| CME100C | Diacylglycerol acyltransferase | 0.7 | 0.8 | 0.8 | 1.4 |
| CMJ162C | Diacylglycerol acyltransferase | 1.1 | 1.4 | 1.5 | 2.1 |
| CMB069C | Diacylglycerol acyltransferase | 1.1 | 1.3 | 2.4 | 1.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takahashi, S.; Okubo, R.; Kanesaki, Y.; Zhou, B.; Takaya, K.; Watanabe, S.; Tanaka, K.; Imamura, S. Identification of Transcription Factors and the Regulatory Genes Involved in Triacylglycerol Accumulation in the Unicellular Red Alga Cyanidioschyzon merolae. Plants 2021, 10, 971. https://doi.org/10.3390/plants10050971
Takahashi S, Okubo R, Kanesaki Y, Zhou B, Takaya K, Watanabe S, Tanaka K, Imamura S. Identification of Transcription Factors and the Regulatory Genes Involved in Triacylglycerol Accumulation in the Unicellular Red Alga Cyanidioschyzon merolae. Plants. 2021; 10(5):971. https://doi.org/10.3390/plants10050971
Chicago/Turabian StyleTakahashi, Sota, Riho Okubo, Yu Kanesaki, Baifeng Zhou, Kazuhiro Takaya, Satoru Watanabe, Kan Tanaka, and Sousuke Imamura. 2021. "Identification of Transcription Factors and the Regulatory Genes Involved in Triacylglycerol Accumulation in the Unicellular Red Alga Cyanidioschyzon merolae" Plants 10, no. 5: 971. https://doi.org/10.3390/plants10050971
APA StyleTakahashi, S., Okubo, R., Kanesaki, Y., Zhou, B., Takaya, K., Watanabe, S., Tanaka, K., & Imamura, S. (2021). Identification of Transcription Factors and the Regulatory Genes Involved in Triacylglycerol Accumulation in the Unicellular Red Alga Cyanidioschyzon merolae. Plants, 10(5), 971. https://doi.org/10.3390/plants10050971