Transgene CpNAC68 from Wintersweet (Chimonanthus praecox) Improves Arabidopsis Survival of Multiple Abiotic Stresses
Abstract
:1. Introduction
2. Results
2.1. Cloning and Sequence Analysis of CpNAC68
2.2. Subcellular Localization and Transcriptional Activation Assay
2.3. Expression Analysis of CpNAC68
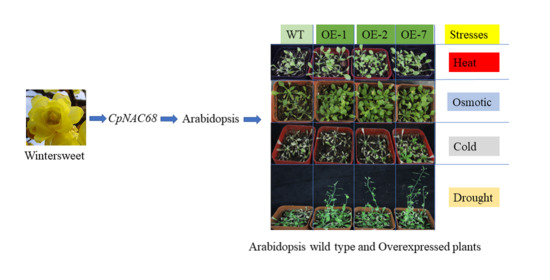
2.4. Abiotic Stress Tolerance of Transgenic Arabidopsis Overexpressing CpNAC68
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Cloning and Sequence Analysis
4.3. CpNAC68 Expression Analyses
4.4. Subcellular Localization and Transactivation Activity Assay of CpNAC68
4.5. Overexpressed Plasmid Construction and Arabidopsis Transformation
4.6. Stress Treatments and Phenotype Observation
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Toth, Z.; Kiss, E.; Kovacs, L. NAC transcription factors as key regulators in stress responses. Acta Hortic. 2014, 1082, 293–298. [Google Scholar] [CrossRef]
- Franco, A.A.; Odom, R.S.; Rando, T.A. Regulation of antioxidant enzyme gene expression in response to oxidative stress and during differentiation of mouse skeletal muscle. Free Radic. Biol. Med. 1999, 27, 1122–1132. [Google Scholar] [CrossRef]
- Tucker, M.R.; Lou, H.; Aubert, M.K.; Wilkinson, L.; Little, A.; Houston, K.; Pinto, S.C.; Shirley, N.J. Exploring the Role of Cell Wall-Related Genes and Polysaccharides during Plant Development. Plants 2018, 7, 42. [Google Scholar] [CrossRef] [Green Version]
- Erpen, L.; Devi, H.S.; Grosser, J.W.; Dutt, M. Potential use of the DREB/ERF, MYB, NAC and WRKY transcription factors to improve abiotic and biotic stress in transgenic plants. Plant Cell Tissue Organ. Cult. (PCTOC) 2018, 132, 1–25. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.-Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis Transcription Factors: Genome-Wide Comparative Analysis Among Eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Dane, F. NAC (NAM/ATAF/CUC) transcription factors in different stresses and their signaling pathway. Acta Physiol. Plant. 2013, 35, 1397–1408. [Google Scholar] [CrossRef]
- Ernst, H.A.; Olsen, A.N.; Skriver, K.; Larsen, S.; Leggio, L.L. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 2004, 5, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Gan, S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006, 46, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Wang, Y.; Zhu, Y.; Tang, J.; Hu, B.; Liu, L.; Ou, S.; Wu, H.; Sun, X.; Chu, J.; et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 10013–10018. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.-Z.; Baig, M.N.R.; Fan, R.; Ye, J.; Cao, Y.-C.; Deng, X.-X. Identification and Expression Pattern of a Novel NAM, ATAF, and CUC-Like Gene from Citrus sinensis Osbeck. Plant Mol. Biol. Rep. 2009, 27, 292–297. [Google Scholar] [CrossRef]
- Fan, J.-P.; Zhao, R. Cloning and Expression Pattern Analysis of the ApNAP Gene in Asarina procumbens. Plant Sci. J. 2014, 32, 251–258. [Google Scholar] [CrossRef]
- Chen, X.; Wang, Y.; Lv, B.; Li, J.; Luo, L.; Lu, S.; Zhang, X.; Ma, H.; Ming, F. The NAC Family Transcription Factor OsNAP Confers Abiotic Stress Response Through the ABA Pathway. Plant Cell Physiol. 2014, 55, 604–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souer, E.; van Houwelingen, A.; Kloos, D.; Mol, J.; Koes, R. The No Apical Meristem Gene of Petunia Is Required for Pattern Formation in Embryos and Flowers and Is Expressed at Meristem and Primordia Boundaries. Cell 1996, 85, 159–170. [Google Scholar] [CrossRef] [Green Version]
- Duval, M.; Hsieh, T.-F.; Kim, S.Y.; Thomas, T.L. Molecular characterization of AtNAM: A member of the Arabidopsis NAC domain superfamily. Plant Mol. Biol. 2002, 50, 237–248. [Google Scholar] [CrossRef]
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 1997, 9, 841–857. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Sun, J.; Xu, P.; Zhang, R.; Li, L. Intron-Mediated Alternative Splicing of WOOD-ASSOCIATED NAC TRANSCRIPTION FACTOR1B Regulates Cell Wall Thickening during Fiber Development in Populus Species. Plant Physiol. 2014, 164, 765–776. [Google Scholar] [CrossRef] [Green Version]
- Hu, R.; Qi, G.; Kong, Y.; Kong, D.; Gao, Q.; Zhou, G. Comprehensive Analysis of NAC Domain Transcription Factor Gene Family in Populus trichocarpa. BMC Plant Biol. 2010, 10, 145. [Google Scholar] [CrossRef] [Green Version]
- Ohashi-Ito, K.; Oda, Y.; Fukuda, H. Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 Directly Regulates the Genes That Govern Programmed Cell Death and Secondary Wall Formation during Xylem Differentiation. Plant Cell 2010, 22, 3461–3473. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, M.; Kubo, M.; Fukuda, H.; Demura, T. VASCULAR-RELATED NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 2008, 55, 652–664. [Google Scholar] [CrossRef] [PubMed]
- Christianson, J.A.; Dennis, E.S.; Llewellyn, D.; Wilson, I. ATAF NAC transcription factors: Regulators of plant stress signaling. Plant Signal. Behav. 2010, 5, 428–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhan, P.-L.; Ke, S.-W.; Zhang, P.-Y.; Zhou, C.-C.; Fu, B.-L.; Zhang, X.-Q.; Zhong, T.-X.; Chen, S.; Xie, X.-M. Overexpression of two cold-responsive ATAF-like NAC transcription factors from fine-stem stylo (Stylosanthes guianensis var. intermedia) enhances cold tolerance in tobacco plants. Plant Cell Tissue Organ. Cult. (PCTOC) 2018, 135, 545–558. [Google Scholar] [CrossRef]
- Gong, L.; Zhang, H.; Liu, X.; Gan, X.; Nie, F.; Yang, W.; Zhang, L.; Chen, Y.; Song, Y.; Zhang, H. Ectopic expression of HaNAC1, an ATAF transcription factor from Haloxylon ammodendron, improves growth and drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2020, 151, 535–544. [Google Scholar] [CrossRef]
- Jia, D.; Gong, X.; Li, M.; Li, C.; Sun, T.; Ma, F. Overexpression of a Novel Apple NAC Transcription Factor Gene, MdNAC1, Confers the Dwarf Phenotype in Transgenic Apple (Malus domestica). Genes 2018, 9, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laubscher, M.; Brown, K.; Tonfack, L.B.; Myburg, A.A.; Mizrachi, E.; Hussey, S.G. Temporal analysis of Arabidopsis genes activated by Eucalyptus grandis NAC transcription factors associated with xylem fibre and vessel development. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Yao, L.; Zhang, N.; Yang, J.; Zhu, X.; Tang, X.; Calderón-Urrea, A.; Si, H. Lateral Root Development in Potato Is Mediated by Stu-mi164 Regulation of NAC Transcription Factor. Front. Plant Sci. 2018, 9, 383. [Google Scholar] [CrossRef]
- Jin, R.; Zhu, Q.-G.; Shen, X.-Y.; Wang, M.-M.; Jamil, W.; Grierson, D.; Yin, X.-R.; Chen, K.-S. DkNAC7, a novel high-CO2/hypoxia-induced NAC transcription factor, regulates persimmon fruit de-astringency. PLoS ONE 2018, 13, e0194326. [Google Scholar] [CrossRef] [Green Version]
- Moyano, E.; Martínez-Rivas, F.J.; Blanco-Portales, R.; Molina-Hidalgo, F.J.; Ric-Varas, P.; Matas-Arroyo, A.J.; Caballero, J.L.; Muñoz-Blanco, J.; Rodríguez-Franco, A. Genome-wide analysis of the NAC transcription factor family and their expression during the development and ripening of the Fragaria × ananassa fruits. PLoS ONE 2018, 13, e0196953. [Google Scholar] [CrossRef] [Green Version]
- Nuruzzaman, M.; Sharoni, A.M.; Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013, 4, 248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eshao, H.-B.; Ewang, H.; Etang, X. NAC transcription factors in plant multiple abiotic stress responses: Progress and prospects. Front. Plant Sci. 2015, 6, 902. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Zhu, L.; Xu, L.; Guo, W.; Zhang, X. GhATAF1, a NAC transcription factor, confers abiotic and biotic stress responses by regulating phytohormonal signaling networks. Plant Cell Rep. 2016, 35, 2167–2179. [Google Scholar] [CrossRef]
- Nagahage, I.S.P.; Sakamoto, S.; Nagano, M.; Ishikawa, T.; Mitsuda, N.; Kawai-Yamada, M.; Yamaguchi, M. An Arabidopsis NAC domain transcription factor, ATAF2, promotes age-dependent and dark-induced leaf senescence. Physiol. Plant. 2020, 170, 299–308. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, C.; Qiu, K.; Chen, H.; Li, Z.; Li, X.; Song, J.; Wang, X.; Gao, J.; Kuai, B.; et al. The transcription factor ZmNAC126 accelerates leaf senescence downstream of the ethylene signalling pathway in maize. Plant Cell Environ. 2020, 43, 2287–2300. [Google Scholar] [CrossRef]
- Huysmans, M.; Buono, R.A.; Skorzinski, N.; Radio, M.C.; De Winter, F.; Parizot, B.; Mertens, J.; Karimi, M.; Fendrych, M.; Nowack, M.K. NAC Transcription Factors ANAC087 and ANAC046 Control Distinct Aspects of Programmed Cell Death in the Arabidopsis Columella and Lateral Root Cap. Plant Cell 2018, 30, 2197–2213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gill, S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Lindemose, S.; Jensen, M.K.; Van De Velde, J.; O’Shea, C.; Heyndrickx, K.S.; Workman, C.; Vandepoele, K.; Skriver, K.; De Masi, F. A DNA-binding-site landscape and regulatory network analysis for NAC transcription factors in Arabidopsis thaliana. Nucleic Acids Res. 2014, 42, 7681–7693. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.F.; Wang, Z.Q.; He, Q.Y.; Wang, J.Y.; Li, P.F.; Xu, J.M.; Zheng, S.J.; Fan, W.; Yang, J.L. Genome-wide identification and expression analysis of the NAC transcription factor family in tomato (Solanum lycopersicum) during aluminum stress. BMC Genom. 2020, 21, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Yao, W.; Li, C.; Lin, S.; Wang, J.; Zhou, B.; Jiang, T. Transcriptome analysis of salt-responsive and wood-associated NACs in Populus simonii × Populus nigra. BMC Plant Biol. 2020, 20, 317. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.-M.; Huang, X.-Q.; Yin, F.-Y.; Li, D.-Q.; Chen, Y.; Chen, L.; Cheng, Z.-Q. Research progress on NAC transcription factor family in Oryza sativa L. Plant Sci. J. 2020, 38, 278–287. [Google Scholar] [CrossRef]
- Saidi, M.N.; Mergby, D.; Brini, F. Identification and expression analysis of the NAC transcription factor family in durum wheat (Triticum turgidum L. ssp. durum). Plant Physiol. Biochem. 2017, 112, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.; Wang, M.; Miao, Y.; Ni, M.; Bibi, N.; Yuan, S.; Li, F.; Wang, X. Molecular Evolution and Expansion Analysis of the NAC Transcription Factor in Zea mays. PLoS ONE 2014, 9, e111837. [Google Scholar] [CrossRef] [Green Version]
- Zhao, K.-G.; Zhou, M.-Q.; Chen, L.-Q.; Zhang, D.; Robert, G.W. Genetic Diversity and Discrimination of Chimonanthus praecox (L.) Link Germplasm Using ISSR and RAPD Markers. HortScience 2007, 42, 1144–1148. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Huang, R.; Ma, J.; Sui, S.; Guo, Y.; Liu, D.; Li, Z.; Lin, Y.; Li, M. Two C3H Type Zinc Finger Protein Genes, CpCZF1 and CpCZF2, from Chimonanthus praecox Affect Stamen Development in Arabidopsis. Genes 2017, 8, 199. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Jiang, Y.; Liu, D.; Ma, J.; Li, J.; Li, M.; Sui, S. Floral Scent Emission from Nectaries in the Adaxial Side of the Innermost and Middle Petals in Chimonanthus praecox. Int. J. Mol. Sci. 2018, 19, 3278. [Google Scholar] [CrossRef] [Green Version]
- Tian, J.; Ma, Z.; Zhao, K.; Zhang, J.; Xiang, L.; Chen, L. Transcriptomic and proteomic approaches to explore the differences in monoterpene and benzenoid biosynthesis between scented and unscented genotypes of wintersweet. Physiol. Plant. 2019, 166, 478–493. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Liu, D.; Ma, J.; Li, M.; Sui, S. CpWRKY71, a WRKY Transcription Factor Gene of Wintersweet (Chimonanthus praecox), Promotes Flowering and Leaf Senescence in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 5325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aslam, M.Z.; Lin, X.; Li, X.; Yang, N.; Chen, L. Molecular Cloning and Functional Characterization of CpMYC2 and CpBHLH13 Transcription Factors from Wintersweet (Chimonanthus praecox L.). Plants 2020, 9, 785. [Google Scholar] [CrossRef]
- Zhao, R.; Song, X.; Yang, N.; Chen, L.; Xiang, L.; Liu, X.-Q.; Zhao, K. Expression of the subgroup IIIf bHLH transcription factor CpbHLH1 from Chimonanthus praecox (L.) in transgenic model plants inhibits anthocyanin accumulation. Plant Cell Rep. 2020, 39, 891–907. [Google Scholar] [CrossRef]
- Liu, D.; Sui, S.; Ma, J.; Li, Z.; Guo, Y.; Luo, D.; Yang, J.; Li, M. Transcriptomic Analysis of Flower Development in Wintersweet (Chimonanthus praecox). PLoS ONE 2014, 9, e86976. [Google Scholar] [CrossRef] [Green Version]
- Mohanta, T.K.; Yadav, D.; Khan, A.; Hashem, A.; Tabassum, B.; Khan, A.L.; Allah, E.F.A.; Al-Harrasi, A. Genomics, molecular and evolutionary perspective of NAC transcription factors. PLoS ONE 2020, 15, e0231425. [Google Scholar] [CrossRef] [Green Version]
- Tran, L.-S.; Nakashima, K.; Sakuma, Y.; Osakabe, Y.; Qin, F.; Simpson, S.D.; Maruyama, K.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J. 2006, 49, 46–63. [Google Scholar] [CrossRef] [PubMed]
- Mitsuda, N.; Matsui, K.; Ikeda, M.; Nakata, M.; Oshima, Y.; Nagatoshi, Y.; Ohme-Takagi, M. CRES-T, An Effective Gene Silencing System Utilizing Chimeric Repressors. Plant Transcr. Factors 2011, 754, 87–105. [Google Scholar]
- Fang, Y.; Liao, K.; Du, H.; Xu, Y.; Song, H.; Li, X.; Xiong, L. A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J. Exp. Bot. 2015, 66, 6803–6817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, Z.; Wang, S.; Zhang, F.; Chen, L.; Hao, X.; Pan, Q.; Fu, X.; Li, L.; Sun, X.; Tang, K. Overexpression of a Novel NAC Domain-Containing Transcription Factor Gene (AaNAC1) Enhances the Content of Artemisinin and Increases Tolerance to Drought and Botrytis cinereal in Artemisia annua. Plant Cell Physiol. 2016, 57, 1961–1971. [Google Scholar] [CrossRef] [Green Version]
- Gunapati, S.; Naresh, R.; Ranjan, S.; Nigam, D.; Hans, A.; Verma, P.C.; Gadre, R.; Pathre, U.V.; Sane, A.P.; Sane, V.A. Expression of GhNAC2 from G. herbaceum, improves root growth and imparts tolerance to drought in transgenic cotton and Arabidopsis. Sci. Rep. 2016, 6, 24978. [Google Scholar] [CrossRef]
- Hou, X.-M.; Zhang, H.-F.; Liu, S.-Y.; Wang, X.-K.; Zhang, Y.-M.; Meng, Y.-C.; Luo, D.; Chen, R.-G. The NAC transcription factor CaNAC064 is a regulator of cold stress tolerance in peppers. Plant Sci. 2020, 291, 110346. [Google Scholar] [CrossRef]
- Jahan, A.; Harris, B.; Lowery, M.; Coburn, K.; Infante, A.M.; Percifield, R.J.; Ammer, A.G.; Kovinich, N. The NAC family transcription factor GmNAC42–1 regulates biosynthesis of the anticancer and neuroprotective glyceollins in soybean. BMC Genom. 2019, 20, 149. [Google Scholar] [CrossRef]
- Yokotani, N.; Ichikawa, T.; Kondou, Y.; Matsui, M.; Hirochika, H.; Iwabuchi, M.; Oda, K. Tolerance to various environmental stresses conferred by the salt-responsive rice gene ONAC063 in transgenic Arabidopsis. Planta 2009, 229, 1065–1075. [Google Scholar] [CrossRef]
- Yokotani, N.; Tsuchida-Mayama, T.; Ichikawa, H.; Mitsuda, N.; Ohme-Takagi, M.; Kaku, H.; Minami, E.; Nishizawa, Y. OsNAC111, a Blast Disease–Responsive Transcription Factor in Rice, Positively Regulates the Expression of Defense-Related Genes. Mol. Plant-Microbe Interact. 2014, 27, 1027–1034. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Wang, J.W.; Sun, L.J.; Song, X.S. The molecular cloning and functional characterization of ChNAC1, a NAC transcription factor in Cerasus humilis. Plant Growth Regul. 2019, 89, 331–343. [Google Scholar] [CrossRef]
- Sun, D.; Zhang, X.; Zhang, Q.; Ji, X.; Jia, Y.; Wang, H.; Niu, L.; Zhang, Y. Comparative transcriptome profiling uncovers a Lilium regale NAC transcription factor, LrNAC35, contributing to defence response against cucumber mosaic virus and tobacco mosaic virus. Mol. Plant Pathol. 2019, 20, 1662–1681. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.; Liu, X.; Niu, F.; Zhao, Q.; Fan, N.; Cao, D.; Meng, D.; He, W.; Guo, B.; Wei, Y.; et al. OoNAC72, a NAC-Type Oxytropis ochrocephala Transcription Factor, Conferring Enhanced Drought and Salt Stress Tolerance in Arabidopsis. Front. Plant Sci. 2019, 10, 890. [Google Scholar] [CrossRef] [Green Version]
- Negi, S.; Tak, H.; Ganapathi, T.R. Expression analysis of MusaNAC68 transcription factor and its functional analysis by overexpression in transgenic banana plants. Plant Cell Tissue Organ. Cult. (PCTOC) 2016, 125, 59–70. [Google Scholar] [CrossRef]
- Mendes, G.C.; Reis, P.A.B.; Calil, I.P.; Carvalho, H.H.; Aragão, F.J.L.; Fontes, E.P.B. GmNAC30 and GmNAC81 integrate the endoplasmic reticulum stress- and osmotic stress-induced cell death responses through a vacuolar processing enzyme. Proc. Natl. Acad. Sci. USA 2013, 110, 19627–19632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ezhang, L.N.; Ezhang, L.C.; Exia, C.; Ezhao, G.; Ejia, J.; Ekong, X.Y. The Novel Wheat Transcription Factor TaNAC47 Enhances Multiple Abiotic Stress Tolerances in Transgenic Plants. Front. Plant Sci. 2016, 6, 1174. [Google Scholar] [CrossRef]
- Negi, S.; Tak, H.; Ganapathi, T.R. Overexpression of MusaNAC68 reduces secondary wall thickness of xylem tissue in banana. Plant Biotechnol. Rep. 2019, 13, 151–160. [Google Scholar] [CrossRef]
- Li, X.-L.; Yang, X.; Hu, Y.-X.; Yu, X.-D.; Li, Q.-L. A novel NAC transcription factor from Suaeda liaotungensis K. enhanced transgenic Arabidopsis drought, salt, and cold stress tolerance. Plant Cell Rep. 2014, 33, 767–778. [Google Scholar] [CrossRef]
- Mao, X.; Chen, S.; Li, A.; Zhai, C.; Jing, R. Novel NAC Transcription Factor TaNAC67 Confers Enhanced Multi-Abiotic Stress Tolerances in Arabidopsis. PLoS ONE 2014, 9, e84359. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Cao, S.; Guan, C.; Kong, X.; Cui, Y.; Liu, B.; Zhou, Y.; Zhang, Y. Overexpressing the NAC transcription factor LpNAC13 from Lilium pumilum in tobacco negatively regulates the drought response and positively regulates the salt response. Plant Physiol. Biochem. 2020, 149, 96–110. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sui, S.; Luo, J.; Ma, J.; Zhu, Q.; Lei, X.; Li, M. Generation and Analysis of Expressed Sequence Tags from Chimonanthus praecox (Wintersweet) Flowers for Discovering Stress-Responsive and Floral Development-Related Genes. Comp. Funct. Genom. 2012, 2012, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clough, S.J.; Bent, A. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dionisio-Sese, M.L.; Tobita, S. Antioxidant responses of rice seedlings to salinity stress. Plant Sci. 1998, 135, 1–9. [Google Scholar] [CrossRef]
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, J.; Liu, D.; Wang, X.; Ahmed, S.; Li, M.; Kovinich, N.; Sui, S. Transgene CpNAC68 from Wintersweet (Chimonanthus praecox) Improves Arabidopsis Survival of Multiple Abiotic Stresses. Plants 2021, 10, 1403. https://doi.org/10.3390/plants10071403
Lin J, Liu D, Wang X, Ahmed S, Li M, Kovinich N, Sui S. Transgene CpNAC68 from Wintersweet (Chimonanthus praecox) Improves Arabidopsis Survival of Multiple Abiotic Stresses. Plants. 2021; 10(7):1403. https://doi.org/10.3390/plants10071403
Chicago/Turabian StyleLin, Jie, Daofeng Liu, Xia Wang, Sajjad Ahmed, Mingyang Li, Nik Kovinich, and Shunzhao Sui. 2021. "Transgene CpNAC68 from Wintersweet (Chimonanthus praecox) Improves Arabidopsis Survival of Multiple Abiotic Stresses" Plants 10, no. 7: 1403. https://doi.org/10.3390/plants10071403
APA StyleLin, J., Liu, D., Wang, X., Ahmed, S., Li, M., Kovinich, N., & Sui, S. (2021). Transgene CpNAC68 from Wintersweet (Chimonanthus praecox) Improves Arabidopsis Survival of Multiple Abiotic Stresses. Plants, 10(7), 1403. https://doi.org/10.3390/plants10071403