1. Introduction
LEAFY (
LFY) and its homologs across species are highly conserved in protein structure and play key roles in coordinating the transition from vegetative to reproductive growth, and in inflorescence patterning.
LFY was initially characterized in
Arabidopsis, where loss-of-function mutants (
lfy) grew shoots or unusual flowers with leaf-like floral organs. Additional experiments demonstrated that
LFY is key for floral meristem identity gene in
Arabidopsis.
LFY is typically present in a single copy but does have two homologs in some lineages (reviewed in [
1,
2]). The origins of
LFY itself appears to pre-date both seed plants and flowering plants as
LFY-like genes are present in moss, ferns, liverworts, and hornworts, as well as plants that form multicellular embryos but not seeds (reviewed in [
2]). In seedless plants, LFY function appears to be key for sporophyte cell division [
2]. In gymnosperms,
LFY and its related gene
NEEDLY (
NLY) are expressed in meristems and appear to be important for both vegetative and reproductive meristem establishment (reviewed in [
2]). The later-evolved primary targets of
LFY in seed and flowering plants, the MADS family, appear to have arisen via MADS family expansion during speciation as there are no known orthologs in bryophytes or ferns [
2,
3,
4,
5].
It appears that some aspects of early
LFY function have been retained in angiosperms outside of floral determination and patterning. For example, many herbaceous flowering plants show low levels of
LFY expression in a range of vegetative tissues.
NFL, the
LFY homolog from
Nicotiana tabacum (tobacco), is expressed in vegetative meristems [
6]. Similarly,
FA, the
Lycopersicon esculentum (tomato) homolog is expressed in vegetative meristems, leaf primordia, and leaves [
7]. In
Fabales (pea),
PEAFLO is an
LFY homolog expressed in leaves [
8]. This trend of
LFY expression beyond floral tissues holds true for the very small number of studies of
LFY in woody perennials. In several species of
Cornus (dogwood) trees, the
LFY homolog is present in leaves and bracts, with an expression pattern that appears to relate to the architecture of the overall inflorescence [
1]. Poplar trees, the focus of this paper, have
LFY expression in floral buds, flowers, vegetative buds, and leaf primordia [
9].
Overexpression of
LFY leads to strong alterations not only in flowers, but also in overall plant architecture. For example, 35S-
NFL1 transformed tobacco plants (tobacco
LFY homolog) grew a single terminal flower and also had reduced stature [
10]. Similarly, expression of 35S-
LFY in hybrid poplar led to a bushy stature and small leaves [
9]. While overexpression analysis of function can create de novo phenotypes it is also a useful means for analyzing gene function. In the case of LFY, overexpression in
Arabidopsis leads to conversion of vegetative meristems to floral meristems and more rapid flowering [
2,
11,
12,
13]. These findings demonstrate the key role of
LFY in floral meristem identity.
Information on functions of
LFY in non-flowering plants is hampered by a paucity of loss-of-function mutants. Studies in
Physcomitrella patens (moss) found that
PpLFY double nulls show normal gametophytes, but the sporophyte either fails to develop after fertilization or has severe growth defects [
14]. These data imply a key role of
LFY in vegetative growth in the diploid life stage. Most loss-of-function studies of
LFY are from examination of flowering plants, and focus on floral alterations, which are striking in nulls. There are very limited studies of the vegetative features of loss-of-function mutants of floral development genes, most studies are in herbaceous annuals, and none analyzed productivity effects in a randomized trial. For example, a novel
lfy mutant in
A. thaliana ecotype No-0 showed increased stem height and additional branches [
15]. Similarly, tomato
fa mutants (loss of
FLORICAULA, tomato homology of
LFY) leads to shoots growing in place of flowers, giving plants a bushy appearance [
7]. Loss of the
LFY homolog from peas gave alterations in leaf shape, with the formation of single rather than compound leaves, which is a dramatic alteration in overall leaf architecture [
8]. Apart from the work of Klocko et al. 2016 [
16], which is closely related to the present study, we are unaware of randomized or field-based studies of vegetative characteristics in
LFY mutants or transgenics.
One potential application of loss-of-function
LFY mutants is for prevention of unwanted gene flow by changing reproductive floral whorls into vegetative structures [
16,
17]. However, this usage would be limited to situations where the desired plant product is a vegetative structure such as a woody stem, and not a seed or fruit. Such applications are mostly likely when trees are grown for bioenergy, pulp, or wood products [
17]. In such cases, it will be essential to determine if loss of
LFY leads to alterations in plant survival, performance, or vegetative traits. Previously, we studied suppression of
LFY in a large field trial of poplar trees [
16], which included many constructs designed to alter the function of a variety of floral genes [
18]. Of these, the strongly suppressed RNAi-
LFY events gave strong alteration in floral form, leading to sterile female flowers, but statistically significant differences in tree growth were not detected. Due to the large-scale nature of the test (many constructs and gene insertion events, only a few replicates per event), and that there were only two
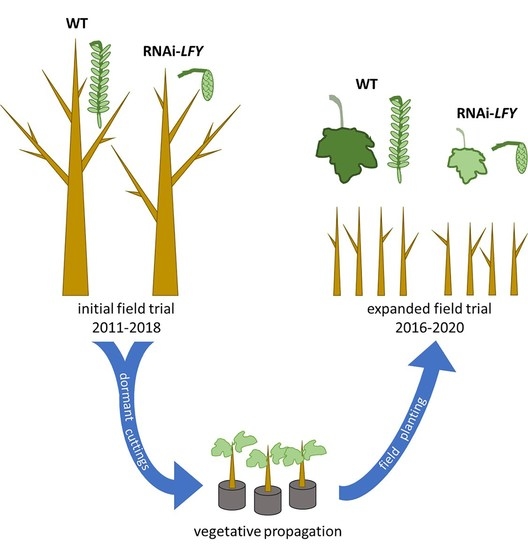
LFY-suppressed sterile events observed, the power of this finding is low. We therefore vegetatively propagated several representative events from the original trial to create a much more highly replicated field trial with more statistical power to detect event differences if they are present. We report that although the trees appeared largely identical in vegetative form, the test had sufficient power to detect some differences in vegetative traits, mainly reductions in productivity traits including stem growth and leaf size that are of great interest for forestry applications.
3. Discussion
An important aspect of this study was to assess the stability of floral phenotypes of sterile RNAi-
LFY trees after propagation through vegetative cuttings. As these trees are female-sterile, they could not be propagated through seeds. Additionally, plantation trees are generally established with cuttings, either rooted or dormant, or via grafted plants, rather than seeds (for example, see [
19,
20,
21]). We noted early on that one event, TN4, performed poorly in both greenhouse and field conditions. In hindsight, this event should have been excluded from the field planting. However, this event showed no indication of such poor performance in the initial field trial testing of RNAi-
LFY events [
16]. It is possible that the cuttings used for propagating these samples were in poor health. Of the 40 initial cuttings for this event, only 10 rooted in greenhouse conditions, and all but 1 of these rooted cuttings were from the same parent tree. It is also possible that there was tree-to-tree variation in rooting ability, as rooting of mature tree cuttings is known to be challenging and variable due to physiological differences in vigor and location/maturity of cuttings [
22].
We found that the use of mature poplar trees for cuttings initially lead to maintained maturity (early flowering), as we observed flowering for three trees in 2017, the first spring after planting. However, all trees reverted to a more juvenile state the next year, with no trees flowering in 2018. The rooted cuttings apparently underwent a developmental reset to reenter the juvenile phase, thus observed phenotypes were not simply due to carryover of parental tree performance. This is commonly seen in poplar plantations, even when large trees or hedges are used as sources of cuttings [
23]. In 2019 most trees of this precocious genotype flowered (91.7%), giving us a good view of floral form, timing, and abundance. Importantly, we found that floral phenotypes were stable between the initial field trial and this new one established with cuttings, with catkins of our two sterile events being smaller than control catkins with no externally visible carpels. Therefore, it is possible to see continued sterility after propagation with RNAi as the molecular approach to achieve gene suppression and genetic containment. As with most other studies of transgenic forest trees, the traits imparted were highly stable under vegetative growth and propagation [
24] including those with a focus in suppression of floral genes such as
AGAMOUS in poplar [
25]. However, to our knowledge, this is the first observation of stability of floral sterility after vegetative propagation.
Analysis of floral form and timing showed that the TS trees showed delayed floral bud burst and small catkins, which is consistent with their initial characterization in the large-scale screening trial. The TS trees also had a lower maximal flowering score (the highest score of open floral buds achieved), and a lower sum of floral scores (the number of open catkins per tree in 2019). However, these two metrics were a bit challenging to assess as the floral buds for the TS trees were both smaller and opened later than those of C and TN trees, and the TS catkins emerged during vegetative bud flush.
These results were supported by the highly significant PC3, which was strongly influenced by floral traits. PC3 was significantly different between TS and control trees, and the main loadings for PC3 were related to leaf weight, petiole ratios, as well as several flowering traits. In general, the TS trees had late floral opening, lower floral abundance, and a change in the relative petiole dimensions as compared to control trees. These first two traits were easy to assess and quantify in the field, while the changes in leaves and petioles were more subtle and were only revealed during data analysis. TS and TN also showed significant differences in PC3, indicative of the differences in their floral abundance and timing of floral opening. This late floral bud opening may indicate a more vegetative-like identity of the floral buds, which would be consistent with a partial conversion of reproductive structures to vegetative structures. These data also show we were able to successfully maintain female tree sterility after vegetative propagation, as discussed above.
We found that block was a significant factor for performance of nearly all vegetative traits. Trees were initially planted in blocks by size (largest trees in block 1, smallest trees in block 4), which probably gave a growth advantage to the larger trees. Additionally, there was variation present from the environment, as this growing site was not uniform soil quality. There were also confounding factors including deer browse, which occurred unevenly across the site prior to mitigation of deer intrusion (browsing was mainly in block 4, which also had the smallest trees at planting). Block adjustments to data allowed for better resolution of differences between tree groups, as can be seen by the clearer separation of TS trees from TN and C in elliptical plots of PC2 (mainly relating to leaf traits) and PC3 (mainly relating to floral timing and abundance).
We did not observe any major alterations in overall tree architecture or form between our RNAi trees and our control trees. Unsurprisingly, this is in contrast to findings in the annual plant
Arabidopsis, where analysis of a novel
lfy mutant showed very noticeable impacts on vegetative features, such as increased number of rosette leaves, additional stem-born branches, and longer stems related to delayed onset and structure of flowers [
15]. Annual
Arabidopsis plants normally grow several terminal floral structures along a main stem, while the
lfy mutants of
Arabidopsis have branches in these equivalent positions [
15]. By contrast, floral development in perennial poplar starts to form floral buds the growing season prior to floral bud flush, and these buds remain dormant over the winter [
26]. Most floral buds occur around the outermost branches of the crown but can be found on the apex of the main shoot as well [
27]. Thus, floral buds are formed in different places on the plant in poplar as compared to
Arabidopsis and poplar are kept in a dormant state until the next growing season. One vegetative difference was the timing of leaf flush at the tops of trees, with TS trees flushing later than control and TN trees, which was a loading of PC5. Given that the floral buds of TS trees also flushed late, it may be that
LFY has a role in the timing of overall bud flush.
We chose to use normal flowering events as a transgenic control for our study as these trees shared a transformation and propagation history with the transgenic sterile trees. These trees were chosen based on morphologically normal catkins in the initial field trial rather than on levels of
LFY suppression as we did not previously characterize expression levels of all RNAi events. We observed significant differences between the overall tree size of some TN and TS events, with event TS1 being significantly shorter in height and smaller in DBH than event TN1. Event TS1 had the smallest height of all events, while TN1 was the largest event based on height, and was larger on average than non-transgenic controls as well. As events represent individual genetic transformation events with the construct insertion into a different place in the genome, it is expected to generate some event-to-event differences. However, these differences occurred both within a normal flowering event (TN1), indicating little impairment of
LFY function, and events where flowers are sterile (TS1), indicating strong impairment of
LFY function. Thus, it seems unlikely these large growth differences are mainly a result of variation in
LFY expression, however, that was not characterized in this study. Moreover, there was wide variation within the wildtype control trees, a likely result of variation due to propagation timing, plant history on the site, and soil fertility. Nevertheless, our ANOVA results showed that the TS group was significantly different than the TN group for several traits, all of them indicating slower growth rate and smaller leaves in TS vs. TN trees. The productivity difference for volume growth exceeded 25%, a very large potential reduction in productivity. This result was also supported by ANOVA for PC1, the “productivity PC” as discussed under results, where its values were highly significantly different for TS vs. TN in the same direction. Significant differences were also observed in petiole traits, which in poplar are linked to tree growth as changes in petiole size is linked to leaf exposure to light [
28]. In poplars, petiole size not only varies between clones, but also within the canopy of one tree, making it a very plastic trait [
28]. It may be that the size differences observed between some TN and TS events could be due to both direct vegetative roles of
LFY, and indirectly due to alterations in light capture ability as influenced by the petiole architecture.
Given the large within group and event-to-event variation, a much larger number of transgenic sterile and normal events would have been desirable for this study; there were just two sterile and three normal RNAi events, and event TN1 tended to have the largest trees, with the tallest height and largest diameter of all events (
Figure 4), perhaps indicating an overall health difference of this event. Unfortunately, only two events with sufficient
LFY suppression for these studies were discovered from our earlier studies [
16] limiting our scope of inference.
In sum, our findings support a role for LFY function in vegetative features and performance of poplar trees, as has been observed for other angiosperms. Despite considerable variation among and within events, the weight of evidence suggests that LFY suppression is not without deleterious vegetative consequences on productivity. Though our results showing stability of floral phenotypes after vegetative propagation are encouraging, the use of LFY suppression for genetic containment may come at a large cost. Before LFY suppression mutation is deployed for genetic containment, additional studies are needed in relevant genotypes and environments to examine if the productivity losses we observed apply, and if so, what their magnitude is. In addition, other approaches to containment that do not appear to have such costs, such as suppression of AGAMOUS, should continue to be pursued.
4. Materials and Methods
4.1. Plasmid Construction and Plant Transformation
In the initial field trial, the RNAi-LFY cassette was generated in the pHannibal vector using a 285-bp fragment of
PtLFY from
Populus trichocarpa using standard organogenic techniques (as described in [
16,
18]). This was inserted into
Populus alba (
P. alba) female 6K10 clones via a binary vector, pART27 using
Agrobacterium-based transformation. Events were rooted and four ramets per event were produced to be grown in the initial field trial. This initial trial was planted in summer 2011 [
18].
4.2. Propagation and Root Induction
Dormant branch cuttings for propagation were collected in March 2016 from the initial 6K10 sterility trial. We selected non-transgenic control trees (C), four RNAi-
LFY events with normal flowers (TN events), and two RNAi-
LFY events with sterile catkins (TS events) for the field trial (
Table S1). Each event was given a simplified number, rather than the longer event name of the initial trial (
Table S1). Two trees from each of these selected events were used as a source of dormant woody branch cuttings for the focused trial. These cuttings were hand collected from the ends of branches and were approximately 1 foot in length. They were placed into plastic bags and stored at 4° Fahrenheit for 10 days before preparing cuttings and inducing rooting. Branch cuttings were trimmed to 10 cm length with the base cut at a 45° angle. After each cutting was trimmed, it was immediately dipped in a 1:5 dilution of Dip ‘N’ Grow rooting hormone and planted in a 3-inch pot filled with moist potting soil. Rooting hormone was freshly diluted for each event. Dormant buds were removed from each cutting, retaining 1–2 vegetative buds at the apex of the cutting. Inverted plastic bags were placed over pots to maintain high humidity.
4.3. Tree Acclimatization
Potted cuttings were moved to a greenhouse and randomized in two blocks. A shade cloth was used to lower the light intensity. Daytime greenhouse temperatures were between 70- and 85-degrees F, and cuttings were provided with constant water from trays under the pots. Bud flush of the cuttings was measured weekly as an indicator of cutting health. After most cuttings had flushed, cuttings were slowly acclimated to the natural humidity by cutting corners off the inverted plastic bags and eventually removing them entirely over the course of about 4 weeks. The first corner was cut off in week 1, the second corner in week 2, then the top of the bag snipped about half open for week 3 and fully open for week 4. After week 4, bags were removed. Flushed cuttings were moved to a lathhouse (outdoor screened in greenhouse) for acclimatization to outdoor conditions prior to planting. Tree height was measured in the lathhouse and used as a guide for planting in field blocks by size.
4.4. Field Design
Following acclimatization in a lath house (open sided greenhouse), rooted cuttings were planted in the field in July of 2016 (
Figure S1). The trial consisted of 4 randomized blocks, with a one-tree border between each block and a one-tree perimeter border, with 9–40 trees per event (
Figure S2,
Table S4). Blocks consisted of rectangular arrangements of 5 × 7 trees with a 1-tree border between and around each block. The total field measured 90 feet (27.4 m) by 120 feet (36.6 m). Trees were planted at seven foot (2.1 m) spacing, their bases were covered with shade cloths and metal tags were placed for identification. Block designation was based on tree size at planting with block 1 being the largest trees and 4 being the smallest trees. Within the blocks, approximately five trees of each event were planted with randomized locations. Border trees were randomly planted using the remaining trees and included trees from each event as well as controls. On the west exterior border of the field, pairs of each event were planted in a non-randomized order as a showcase for demonstrative purposes. The border trees and showcase trees were excluded from statistical analyses and results. A total of 221 trees were planted: 30 controls, 114 transgenic normal, and 77 transgenic sterile (
Table S4).
This trial was established under permit from APHIS (BRS permit number 17-012-101r-a2, previous permit numbers 17-012-101r-a1, 17-012-101r, 13-330-102r and 10-260-102-a1). Prior to field planting in July 2016, the field was sprayed with herbicide, tilled, and irrigation lines were established. Sprinkler heads with 360° rotation were attached to the irrigation lines which were spread out in an E-shape to provide full coverage of the field trial. From July through August 2016, the field was watered for four hours each morning. In September 2016, fields were watered for two hours each morning. Trees were irrigated according to this plan in 2017 as well, then the irrigation system was removed in early 2018.
4.5. Vegetative Growth Measurements and Spring Leaf Flush Scoring
Tree height was measured at planting and again in each dormant season. Height was collected using a meter stick in the first season of growth and using cm tape in later growth seasons. Stem diameter was measured at 6 inches above soil level using Meba IP54 Electronic Digital Calipers. Volume index was calculated using height × DBH2. Spring leaf flush was scored for the top and bottom half of each tree using the following scoring system; 0 = no leaves flushed, 1 = buds starting to break, 2 = partial leaf emergence, 3 = leaves fully flushed. Tree survival was scored each spring along with the timing of leaf flush. Trees that did not flush leaves were scored as dead. Because event TN4 had poor performance in terms of growth and survival in the field, it was excluded from future analysis.
4.6. Leaf Collection
Each year, three leaves were collected and measured from each tree. Either in the field, or shortly after collection, chlorophyll content was measured using a hand-held SPAD 502 Plus Chlorophyll Meter. Three SPAD measurements taken and averaged on each leaf. During the first growth season, leaves were collected at the visual midpoint of the trees. For subsequent years, leaves were collected using a leaf plasticron index. This was measured by counting ten leaves down from the top of the tree without including nascent leaves. Nascent leaves were those under 4 cm in length. Leaves were collected from different sides of the tree when possible. Any trees with fewer than 10 developed leaves were not included in the statistical analyses. No chlorophyll content was measured in 2019.
4.7. Leaf Trait Measurements
Petiole length and width of fresh leaves were measured with digital calipers. Leaves were scanned using an HP Scanjet 8200. Each scan included three leaves from the same tree and a ruler for a scale marker. Leaf area in cm2 was calculated using ImageJ software. Scanned leaves were placed in drying ovens until the mass of the combined leaves remained stable, indicating leaves were fully dry. Each leaf was weighed with a Mettler AJ100 scale. Leaf-specific weight was calculated as the dry weight divided by the leaf area.
4.8. Scoring Flowering, Floral Timing and Floral Abundance
Trees were scored yearly for the presence of floral buds and fully flushed catkins. Trees with at least one floral bud were scored as flowering. In 2019 trees were scored for floral bud presence (flowering or not), timing of floral opening, and abundance of open flowers (sum of flowering scores). Floral opening was scored for 7 weeks beginning 11 March 2019 using the following system.
- 0.
No fully flushed catkins.
- 1.
Very sparse fully flushed catkins (less than 10).
- 2.
Sparse fully flushed catkins (more than 10 but less than 20).
- 3.
Some fully flushed catkins (more than 20 but less than 40).
- 4.
Many fully flushed catkins (more than 40 but less than 50).
- 5.
Abundant fully flushed catkins (50 or more).
Trees with numbers of fully flushed catkins that fell about halfway between two categories were scored at half integer intervals. Maximal flowering score was the highest overall score given to a tree across the 7 weeks.
Floral abundance was determined by counting the actual number of fully flushed catkins on each tree. This was performed twice, once on 25 March 2019, and once on April 4 2019. These data were termed “sum of floral scores.”
4.9. Field Management
In summer 2016 several of the trees experienced browsing damage from deer. Due to uneven tree performance in the 2016 growing season, it was decided to trim all trees to a uniform height of 30 cm from soil level. Trees that had not reached 30 cm in height were left untrimmed. Trees were trimmed during the first winter season then singled in spring 2017. Trees were terminated in fall 2020 by coppicing (cutting off near ground level), followed by injecting EZ-Ject copperhead herbicide shells containing imazapyr directly into the stem roughly 3 inches (7.6 cm) off the ground. Stems were incinerated while the stumps remained in the ground.
4.10. Data Analysis
All statistical analyses were performed in R studio, raw and analyzed data can be found in Ap raw R code can be found in
Supplementary File S2. All trees that were recorded as dead in any year, even if they resprouted in another year, were excluded from the analysis due to poor growth, as were all data collected in 2016 due to deer damage and trimming.
Using all collected data from 2017–2019, a principal component analysis was performed using the function prcompfrom the stats package [
29]. Prior to analysis, all rows containing missing data were omitted with the function na.omit from the stats package, and all data were adjusted for block. After adjusting for block, the polarity of PCA results changed, meaning the major growth traits are negative, and the more negative a trait is, the larger the tree is in reality. Package dyplr [
30] was used to assign groups and events to their respective blocks using the function mutate. Linear models and ANOVAs were created with the principal components 1–5. All linear models were created using the function lm in the stats package. Linear models for the principal components were created with event as the independent variable.
Traits were independently analyzed within the 2019 collected data only. This is because after the third year of growth, the trees reached maturity and produced mature leaves and flowers. As such, data collected in 2019 were considered the most reliable data. The 2019 data were subset into 4 datasets to exclude missing data values without excluding whole rows. This was done using the function subset in the base R package [
29]. All individual trait analysis was performed on raw data unadjusted for block. Linear models were created for each numerical variable within the 2019 collected data. All linear models were created with event and block as the independent variable. In all linear models, block × event interactions were ignored as they were found to only be significant in 2 variables.
Logistic regressions were created with event as the independent variable for each categorical variable within this dataset with the function glm in the package brglm2 [
31]. Prior to analysis, categorical variables were collapsed into binary variables. All floral scores were collapsed from 0–2 (to become zero) and 3–5 (to become 1), while leaf flush scores were collapsed in the same manner from 0–1 and 2–3. The sum of floral scores over the whole season were collapsed from values greater than the median. All logistic regressions were tested for complete separation. Where complete separation was detected, penalized logistic regressions were created. Diagnostic Q-Q and residual plots created with package DHARMa [
32] and function simulateResiduals indicated that assumptions of normality and equal variance were acceptable. Likelihood ratio tests were performed on all logistic regressions using the function drop1 from brglm2. Manual chi-squared tests were used to uncover differences between classes.
Diagnostic plots were created for all linear models, which found the assumptions of normality, equal variance, and independence to be acceptable. ANOVAs were then performed on all linear models using the function anova in the stats package. These were used to then perform Tukey’s HSD test as well as contrasts using the function emmeans in the package emmeans [
33].
Graphics for all data were created with ggplot2 [
34], ggbiplot [
35] (dependency devtools, ggplot2), plotly [
36], graphics in base R, and multcompView [
37]. The color palette for all graphics is as follows: grey27, royal-blue, and tomato3. All boxplots display block adjusted data.