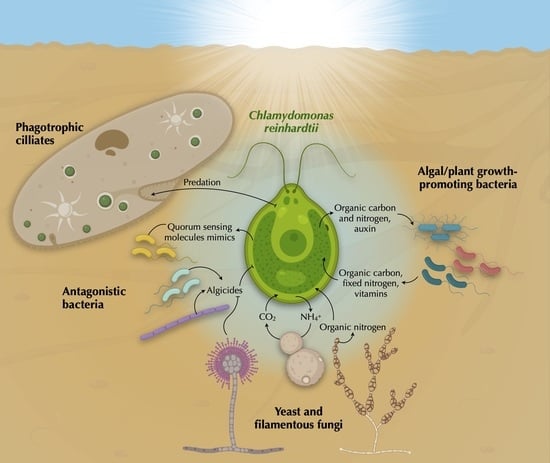
Chlamydomonas reinhardtii, a Reference Organism to Study Algal–Microbial Interactions: Why Can’t They Be Friends?
Abstract
:1. Why Microalgae and Why Chlamydomonas reinhardtii?
2. Biodiversity in the Chlamydomonas Phycosphere
3. Chlamydomonas’ Predators
4. Chlamydomonas’ Fungal Partners
5. Chlamydomonas’ Bacterial Partners
5.1. Vitamin B12 Production
5.2. Nitrogen Fixation
5.3. Amino Acids and Peptides Mineralization
5.4. Auxin Production and Degradation
5.5. Quorum Quenching

6. Harnessing Chlamydomonas—Microbial Interactions for Biotechnological Applications
7. Insights beyond Symbiosis
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gerotto, C.; Norici, A.; Giordano, M. Toward Enhanced Fixation of CO2 in Aquatic Biomass: Focus on Microalgae. Front. Energy Res. 2020, 8, 213. [Google Scholar] [CrossRef]
- Falkowski, P.G. The Role of Phytoplankton Photosynthesis in Global Biogeochemical Cycles. Photosynth. Res. 1994, 39, 235–258. [Google Scholar] [CrossRef] [PubMed]
- Kazmi, S.S.U.H.; Yapa, N.; Karunarathna, S.C.; Suwannarach, N. Perceived Intensification in Harmful Algal Blooms Is a Wave of Cumulative Threat to the Aquatic Ecosystems. Biology 2022, 11, 852. [Google Scholar] [CrossRef]
- Oliveira, C.Y.B.; Jacob, A.; Nader, C.; Oliveira, C.D.L.; Matos, Â.P.; Araújo, E.S.; Shabnam, N.; Ashok, B.; Gálvez, A.O. An Overview on Microalgae as Renewable Resources for Meeting Sustainable Development Goals. J. Environ. Manag. 2022, 320, 115897. [Google Scholar] [CrossRef]
- Sibbald, S.J.; Archibald, J.M. Genomic Insights into Plastid Evolution. Genome Biol. Evol. 2020, 12, 978–990. [Google Scholar] [CrossRef] [PubMed]
- Ramanan, R.; Kim, B.-H.; Cho, D.-H.; Oh, H.-M.; Kim, H.-S. Algae-Bacteria Interactions: Evolution, Ecology and Emerging Applications. Biotechnol. Adv. 2016, 34, 14–29. [Google Scholar] [CrossRef]
- Ray, A.; Nayak, M.; Ghosh, A. A Review on Co-Culturing of Microalgae: A Greener Strategy towards Sustainable Biofuels Production. Sci. Total Environ. 2022, 802, 149765. [Google Scholar] [CrossRef]
- Croft, M.T.; Lawrence, A.D.; Raux-Deery, E.; Warren, M.J.; Smith, A.G. Algae Acquire Vitamin B12 through a Symbiotic Relationship with Bacteria. Nature 2005, 438, 90–93. [Google Scholar] [CrossRef]
- Smith, M.J.; Francis, M.B. A Designed A. vinelandii-S. elongatus Coculture for Chemical Photoproduction from Air, Water, Phosphate, and Trace Metals. ACS Synth. Biol. 2016, 5, 955–961. [Google Scholar] [CrossRef] [PubMed]
- Venkataram, S.; Kuo, H.-Y.; Hom, E.F.Y.; Kryazhimskiy, S. Mutualism-Enhancing Mutations Dominate Early Adaptation in a Two-Species Microbial Community. Nat. Ecol. Evol. 2023, 7, 143–154. [Google Scholar] [CrossRef]
- Kazamia, E.; Helliwell, K.E.; Purton, S.; Smith, A.G.; Fussmann, G. How Mutualisms Arise in Phytoplankton Communities: Building Eco-Evolutionary Principles for Aquatic Microbes. Ecol. Lett. 2016, 19, 810–822. [Google Scholar] [CrossRef]
- Harris, E.H. Introduction into Chlamydomonas and Its Laboratory Use. In The Chlamydomonas Sourcebook; Oxford Academic Press: Oxford, UK, 2009. [Google Scholar]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Maréchal-Drouard, L.; et al. The Chlamydomonas Genome Reveals the Evolution of Key Animal and Plant Functions. Science 2007, 318, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Sasso, S.; Stibor, H.; Mittag, M.; Grossman, A.R. From Molecular Manipulation of Domesticated Chlamydomonas reinhardtii to Survival in Nature. eLife 2018, 7, e39233. [Google Scholar] [CrossRef]
- Salomé, P.A.; Merchant, S.S. A Series of Fortunate Events: Introducing Chlamydomonas as a Reference Organism. Plant Cell 2019, 31, 1682–1707. [Google Scholar] [CrossRef]
- Scaife, M.A.; Nguyen, G.T.D.T.; Rico, J.; Lambert, D.; Helliwell, K.E.; Smith, A.G. Establishing Chlamydomonas reinhardtii as an Industrial Biotechnology Host. Plant J. 2015, 82, 532–546. [Google Scholar] [CrossRef] [PubMed]
- Kelterborn, S.; Boehning, F.; Sizova, I.; Baidukova, O.; Evers, H.; Hegemann, P. Gene Editing in Green Alga Chlamydomonas reinhardtii via CRISPR-Cas9 Ribonucleoproteins. Plant Synth. Biol. Methods Mol. Biol. 2022, 2379, 45–65. [Google Scholar]
- Bell, W.; Mitchell, R. Chemotactic And Growth Responses Of Marine Bacteria To Algal Extracellular Products. Biol. Bull. 1972, 143, 265–277. [Google Scholar] [CrossRef]
- Seymour, J.R.; Amin, S.A.; Raina, J.-B.; Stocker, R. Zooming in on the Phycosphere: The Ecological Interface for Phytoplankton–Bacteria Relationships. Nat. Microbiol. 2017, 2, 17065. [Google Scholar] [CrossRef]
- Ling, N.; Wang, T.; Kuzyakov, Y. Rhizosphere Bacteriome Structure and Functions. Nat. Commun. 2022, 13, 836. [Google Scholar] [CrossRef]
- Cooper, M.B.; Smith, A.G. Exploring Mutualistic Interactions between Microalgae and Bacteria in the Omics Age. Curr. Opin. Plant Biol. 2015, 26, 147–153. [Google Scholar] [CrossRef]
- Krohn-Molt, I.; Alawi, M.; Förstner, K.U.; Wiegandt, A.; Burkhardt, L.; Indenbirken, D.; Thieß, M.; Grundhoff, A.; Kehr, J.; Tholey, A.; et al. Insights into Microalga and Bacteria Interactions of Selected Phycosphere Biofilms Using Metagenomic, Transcriptomic, and Proteomic Approaches. Front. Microbiol. 2017, 8, 1941. [Google Scholar] [CrossRef] [Green Version]
- Durán, P.; Flores-Uribe, J.; Wippel, K.; Zhang, P.; Guan, R.; Melkonian, B.; Melkonian, M.; Garrido-Oter, R. Shared Features and Reciprocal Complementation of the Chlamydomonas and Arabidopsis Microbiota. Nat. Commun. 2022, 13, 406. [Google Scholar] [CrossRef]
- Kim, B.H.; Ramanan, R.; Cho, D.H.; Oh, H.M.; Kim, H.S. Role of Rhizobium, a Plant Growth Promoting Bacterium, in Enhancing Algal Biomass through Mutualistic Interaction. Biomass Bioenergy 2014, 69, 95–105. [Google Scholar] [CrossRef]
- de Jesus Astacio, L.M.; Prabhakara, K.H.; Li, Z.; Mickalide, H.; Kuehn, S. Closed Microbial Communities Self-Organize to Persistently Cycle Carbon. Ecol. Biophys. Comput. Biol. 2021, 118, 2013564118. [Google Scholar] [CrossRef] [PubMed]
- Hekstra, D.R.; Leibler, S. Contingency and Statistical Laws in Replicate Microbial Closed Ecosystems. Cell 2012, 149, 1164–1173. [Google Scholar] [CrossRef]
- Frentz, Z.; Kuehn, S.; Leibler, S. Strongly Deterministic Population Dynamics in Closed Microbial Communities. Phys. Rev. X 2015, 5, 041014. [Google Scholar] [CrossRef]
- Mickalide, H.; Kuehn, S. Higher-Order Interaction between Species Inhibits Bacterial Invasion of a Phototroph-Predator Microbial Community. Cell Syst. 2019, 9, 521–533. [Google Scholar] [CrossRef] [PubMed]
- Lurling, M.; Beekman, W. Palmelloids Formation in Chlamydomonas reinhardtii: Defence against Rotifer Predators? Ann. Limnol. 2006, 42, 65–72. [Google Scholar] [CrossRef]
- Van Donk, E.; Liirling, M.; Hessen, D.O.; Lokhorst, G.M. Altered Cell Wall Morphology in Nutrient-Deficient Phytoplankton and Its Impact on Grazers. Limnol. Ocean. 1997, 42, 357–364. [Google Scholar] [CrossRef]
- Gophen, M. Feeding of Daphnia on Chlamydomonas and Chlorobium. Nature 1977, 265, 271–273. [Google Scholar] [CrossRef]
- Taub, F.B.; Mckenzie, D.H. Continuous Cultures of an Alga and Its Grazer. Bull. Ecol. Res. Comm. 1973, 17, 371–377. [Google Scholar]
- Krespach, M.K.C.; García-Altares, M.; Flak, M.; Schoeler, H.; Scherlach, K.; Netzker, T.; Schmalzl, A.; Mattern, D.J.; Schroeckh, V.; Komor, A.; et al. Lichen-like Association of Chlamydomonas reinhardtii and Aspergillus nidulans Protects Algal Cells from Bacteria. ISME J. 2020, 14, 2794–2805. [Google Scholar] [CrossRef] [PubMed]
- Aiyar, P.; Schaeme, D.; García-Altares, M.; Flores, D.C.; Dathe, H.; Hertweck, C.; Sasso, S.; Mittag, M. Antagonistic Bacteria Disrupt Calcium Homeostasis and Immobilize Algal Cells. Nat. Commun. 2017, 8, 1756. [Google Scholar] [CrossRef] [PubMed]
- Rose, M.M.; Scheer, D.; Hou, Y.; Hotter, V.S.; Komor, A.J.; Aiyar, P.; Scherlach, K.; Vergara, F.; Yan, Q.; Loper, J.E.; et al. The Bacterium Pseudomonas protegens Antagonizes the Microalga Chlamydomonas reinhardtii Using a Blend of Toxins. Environ. Microbiol. 2021, 23, 5525–5540. [Google Scholar] [CrossRef]
- Hotter, V.; Zopf, D.; Joong Kim, H.; Silge, A.; Schmitt, M.; Aiyar, P.; Fleck, J.; Matthäus, C.; Hniopek, J.; Yan, Q.; et al. A Polyyne Toxin Produced by an Antagonistic Bacterium Blinds and Lyses a Chlamydomonas Alga. Proc. Natl. Acad. Sci. USA 2021, 118, 2107695118. [Google Scholar] [CrossRef]
- Iwai, S.; Fujita, K.; Takanishi, Y.; Fukushi, K. Photosynthetic Endosymbionts Benefit from Host’s Phagotrophy, Including Predation on Potential Competitors. Curr. Biol. 2019, 29, 3114–3119. [Google Scholar] [CrossRef]
- Iwasa, K.; Murakami, S. Palmelloid Formation of Chlamydomonas I. Palmelloid Induction by Organic Acids. Physiol. Plantarum 1968, 21, 1224–1233. [Google Scholar] [CrossRef]
- de Carpentier, F.; Lemaire, S.D.; Danon, A. When Unity Is Strength: The Strategies Used by Chlamydomonas to Survive Environmental Stresses. Cells 2019, 8, 1307. [Google Scholar] [CrossRef]
- Herron, M.D.; Borin, J.M.; Boswell, J.C.; Walker, J.; Chen, I.-C.K.; Knox, C.A.; Boyd, M.; Rosenzweig, F.; Ratcliff, W.C. De Novo Origins of Multicellularity in Response to Predation. Sci. Rep. 2019, 9, 2328. [Google Scholar] [CrossRef]
- Bernardes, J.P.; John, U.; Woltermann, N.; Valiadi, M.; Hermann, R.J.; Becks, L.; Wegener, A. The Evolution of Convex Trade-Offs Enables the Transition towards Multicellularity. Nat. Comun. 2021, 12, 4222. [Google Scholar] [CrossRef] [PubMed]
- Becks, L.; Ellner, S.P.; Jones, L.E.; Hairston, N.G. Reduction of Adaptive Genetic Diversity Radically Alters Eco-Evolutionary Community Dynamics. Ecol. Lett. 2010, 13, 980–997. [Google Scholar] [CrossRef] [PubMed]
- Sathe, S.; Durand, P.M. Cellular Aggregation in Chlamydomonas (Chlorophyceae) Is Chimaeric and Depends Ontraits like Cell Size and Motility. Eur. J. Phycol. 2016, 51, 129–138. [Google Scholar] [CrossRef] [Green Version]
- Latta, L.C.; O’donnell, R.P.; Pfrender, M.E.; O’Donnell, R.P.; Pfrender, M.E. Vertical Distribution of Chlamydomonas Changes in Response to Grazer and Predator Kairomones. Oikos 2009, 118, 853–858. [Google Scholar] [CrossRef]
- Grimm, M.; Grube, M.; Schiefelbein, U.; Zühlke, D.; Bernhardt, J.; Riedel, K. The Lichens’ Microbiota, Still a Mystery? Front. Microbiol. 2021, 12, 714. [Google Scholar] [CrossRef]
- Hom, E.F.Y.; Murray, A.W. Niche Engineering Demonstrates a Latent Capacity for Fungal-Algal Mutualism. Science 2014, 345, 94–98. [Google Scholar] [CrossRef]
- Simon, J.; Kósa, A.; Bóka, K.; Vági, P.; Simon-Sarkadi, L.; Mednyánszky, Z.; Horváth, A.N.; Nyitrai, P.; Böddi, B.; Preininger, E. Self-Supporting Artificial System of the Green Alga Chlamydomonas reinhardtii and the Ascomycetous Fungus Alternaria infectoria. Symbiosis 2017, 71, 199–209. [Google Scholar] [CrossRef]
- Sokolovskaya, O.M.; Shelton, A.N.; Taga, M.E. Sharing Vitamins: Cobamides Unveil Microbial Interactions. Science 2020, 369, eaba0165. [Google Scholar] [CrossRef] [PubMed]
- Nef, C.; Dittami, S.; Kaas, R.; Briand, E.; Noël, C.; Mairet, F.; Garnier, M. Sharing Vitamin B12 between Bacteria and Microalgae Does Not Systematically Occur: Case Study of the Haptophyte Tisochrysis lutea. Microorganisms 2022, 10, 1337. [Google Scholar] [CrossRef]
- Bunbury, F.; Deery, E.; Sayer, A.P.; Bhardwaj, V.; Harrison, E.L.; Warren, M.J.; Smith, A.G. Exploring the Onset of B12-Based Mutualisms Using a Recently Evolved Chlamydomonas Auxotroph and B 12-Producing Bacteria. Environ. Microbiol. 2022, 24, 3134–3147. [Google Scholar] [CrossRef]
- Grant, M.A.A.; Kazamia, E.; Cicuta, P.; Smith, A.G. Direct Exchange of Vitamin B12 Is Demonstrated by Modelling the Growth Dynamics of Algal–Bacterial Cocultures. ISME J. 2014, 8, 1418–1427. [Google Scholar] [CrossRef]
- Helliwell, K.E.; Collins, S.; Kazamia, E.; Purton, S.; Wheeler, G.L.; Smith, A.G. Fundamental Shift in Vitamin B12 Eco-Physiology of a Model Alga Demonstrated by Experimental Evolution. ISME J. 2015, 9, 1446–1455. [Google Scholar] [CrossRef]
- Xie, B.; Bishop, S.; Stessman, D.; Wright, D.; Spalding, M.H.; Halverson, L.J. Chlamydomonas reinhardtii Thermal Tolerance Enhancement Mediated by a Mutualistic Interaction with Vitamin B12-Producing Bacteria. ISME J. 2013, 7, 1544–1555. [Google Scholar] [CrossRef]
- Helliwell, K.E.; Wheeler, G.L.; Leptos, K.C.; Goldstein, R.E.; Smith, A.G. Insights into the Evolution of Vitamin B12 Auxotrophy from Sequenced Algal Genomes. Mol. Biol. Evol. 2011, 28, 2921–2933. [Google Scholar] [CrossRef] [PubMed]
- Ellis, K.A.; Cohen, N.R.; Moreno, C.; Marchetti, A. Cobalamin-Independent Methionine Synthase Distribution and Influence on Vitamin B12 Growth Requirements in Marine Diatoms. Protist 2017, 168, 32–47. [Google Scholar] [CrossRef] [PubMed]
- Kazamia, E.; Czesnick, H.; Thanh, T.; Nguyen, V.; Croft, M.T.; Sherwood, E.; Sasso, S.; Hodson, S.J.; Warren, M.J.; Smith, A.G. Mutualistic Interactions between Vitamin B12-Dependent Algae and Heterotrophic Bacteria Exhibit Regulation. Environ. Microbiol. 2012, 14, 1466–1476. [Google Scholar] [CrossRef] [PubMed]
- Helliwell, K.E.; Pandhal, J.; Cooper, M.B.; Longworth, J.; Kudahl, U.J.; Russo, D.A.; Tomsett, E.V.; Bunbury, F.; Salmon, D.L.; Smirnoff, N.; et al. Quantitative Proteomics of a B12-Dependent Alga Grown in Coculture with Bacteria Reveals Metabolic Tradeoffs Required for Mutualism. New Phytol. 2018, 217, 599–612. [Google Scholar] [CrossRef]
- Laeverenz, H.; Id, S.; Peaudecerf Id, J.; Bunbury, F.; Whitehouse, M.J.; Foster, R.A.; Smith, A.G.; Crozeid, O.A. Combining SIMS and Mechanistic Modelling to Reveal Nutrient Kinetics in an Algal-Bacterial Mutualism. PLoS ONE 2021, 16, e0251643. [Google Scholar] [CrossRef]
- Peaudecerf, F.J.; Bunbury, F.; Bhardwaj, V.; Bees, M.A.; Smith, A.G.; Goldstein, R.E.; Croze, O.A. Microbial Mutualism at a Distance: The Role of Geometry in Diffusive Exchanges. Phys. Rev. E 2018, 97, 022411. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.J.; Mens, C.; Hastwell, A.H.; Zhang, M.; Su, H.; Jones, C.H.; Chu, X.; Gresshoff, P.M. Legume Nodulation: The Host Controls the Party. Plant Cell Environ. 2019, 42, 41–51. [Google Scholar] [CrossRef]
- Caputo, A.; Nylander, J.A.A.; Foster, R.A. The Genetic Diversity and Evolution of Diatom-Diazotroph Associations Highlights Traits Favoring Symbiont Integration. FEMS Microbiol. Lett. 2019, 366, fny297. [Google Scholar] [CrossRef]
- Schvarcz, C.R.; Wilson, S.T.; Caffin, M.; Stancheva, R.; Li, Q.; Turk-Kubo, K.A.; White, A.E.; Karl, D.M.; Zehr, J.P.; Steward, G.F. Overlooked and Widespread Pennate Diatom-Diazotroph Symbioses in the Sea. Nat. Commun. 2022, 13, 799. [Google Scholar] [CrossRef]
- Turk-Kubo, K.A.; Mills, M.M.; Arrigo, K.R.; van Dijken, G.; Henke, B.A.; Stewart, B.; Wilson, S.T.; Zehr, J.P. UCYN-A/Haptophyte Symbioses Dominate N2 Fixation in the Southern California Current System. ISME Commun. 2021, 1, 42. [Google Scholar] [CrossRef] [PubMed]
- Villa, J.A.; Ray, E.E.; Barney, B.M. Azotobacter vinelandii Siderophore Can Provide Nitrogen to Support the Culture of the Green Algae Neochloris oleoabundans and Scenedesmus sp. BA032. FEMS Microbiol. Lett. 2014, 351, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Preininger, E.; Ponyi, T.; Sarkadi, L.; Nyitrai, P.; Gyurjan, I. Long-Living Azotobacter-Chlamydomonas Association as a Model System for Plant-Microbe Interactions. Symbiosis 2006, 42, 45–50. [Google Scholar]
- Romano, I.; Ventorino, V.; Pepe, O. Effectiveness of Plant Beneficial Microbes: Overview of the Methodological Approaches for the Assessment of Root Colonization and Persistence. Front. Plant Sci. 2020, 11, 6. [Google Scholar] [CrossRef] [PubMed]
- Gyurjan, I.; Turtoczky, I.; Toth, G.; Paless, G.; Nghia, N.H. Intercellular Symbiosis of Nitrogen-Fixing Bacteria and Green Alga. Acta Bot. Hung 1984, 30, 249–256. [Google Scholar]
- Gyurjan, I.; Nghia, N.H.; Tóth, G.; Turtozky, I.; Stefanovits, P. Photosynthesis, Nitrogen Fixation and Enzyme Activities in Chlamydomonas-Azotobacter Symbioses. Biochem. Physiol. Pflanz. 1986, 181, 147–153. [Google Scholar] [CrossRef]
- Nghia, N.H.; Gyurján, I.; Stefanovits, P.; Turtóczky, I. Establishment of Nitrogen-Fixing Chlamydomonas-Azotobacter Symbioses: Physiological, Biochemical and Morphological Examinations. Endocytobiosis Cell Res. 1986, 3, 179–188. [Google Scholar]
- Koranyi, P.; Gyurjan, I.; Stefanovits, P.; Boka, K.; Szigeti, Z. Characterisation of an Artificial Nitrogen-Fixing Alga-Bacterium Ectocytobiosis. Symbiosis 1990, 8, 175–187. [Google Scholar]
- Nghia, N.H.; Gyurján, I.; Stefanovits, P.; Paless, G.; Turtóczky, I. Uptake of Azotobacters by Somatic Fusion of Cell-Wall Mutants of Chlamydomonas reinhardtii. Biochem. Physiol. Pflanz. 1986, 181, 347–357. [Google Scholar] [CrossRef]
- Gyurjan, I.; Koranyi, P.; Paless, G.Y. Ultrastructural Analysis of an Artificial Alga-Bacterium Endosymbiosis After Prolonged Cultivation. Symbiosis 1992, 14, 475–484. [Google Scholar]
- Calatrava, V.; Hom, E.F.Y.; Llamas, Á.; Fernández, E.; Galván, A. Nitrogen Scavenging from Amino Acids and Peptides in the Model Alga Chlamydomonas reinhardtii. The Role of Extracellular L-Amino Oxidase. Algal Res. 2019, 38, 101395. [Google Scholar] [CrossRef]
- Allen, M.B. Excretion of Organic Compounds by Chlamydomonas. Arch. Mikrobiol. 1956, 24, 163–168. [Google Scholar] [CrossRef]
- Quiroz-Rocha, E.; Moreno, R.; Hernández-Ortíz, A.; Carlos Fragoso-Jiménez, J.; Felipe Muriel-Millán, L.; Guzmán, J.; Espín, G.; Rojo, F.; Núñez, C. Glucose Uptake in Azotobacter vinelandii Occurs through a GluP Transporter That Is under the Control of the CbrA/CbrB and Hfq-Crc Systems. Sci. Rep. 2001, 7, 858. [Google Scholar] [CrossRef]
- Collins, R.P.A.K.K. Keto Acids Produced by Chlamydomonas reinhardtii. Can. J. Microbiol. 1967, 13, 995–999. [Google Scholar] [CrossRef] [PubMed]
- Vogel, S.L.; Frisch, H.L.; Gotham, I.J. Qualitative Assay of Dissolved Amino Acids and Sugars Excreted by Chlamydomonas reinhardtii (Chlorophyceae) and Euglena gracilis (Euglenophyceae). J. Phycol. 1978, 14, 403–406. [Google Scholar] [CrossRef]
- Mus, F.; Dubini, A.; Seibert, M.; Posewitz, M.C.; Grossman, A.R. Anaerobic Acclimation in Chlamydomonas reinhardtii. J. Biol. Chem. 2007, 282, 25475–25486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Heijden, M.G.A.; De Bruin, S.; Luckerhoff, L.; van Logtestijn, R.S.P.; Schlaeppi, K. A Widespread Plant-Fungal-Bacterial Symbiosis Promotes Plant Biodiversity, Plant Nutrition and Seedling Recruitment. ISME J. 2016, 10, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Lorincz, Z.; Preininger, E.; Kósa, A.; Pónyi, T.; Nyitrai, P.; Sarkadi, L.; Kovács, G.M.; Böddi, B.; Gyurján, I. Artificial Tripartite Symbiosis Involving a Green Alga (Chlamydomonas), a Bacterium (Azotobacter) and a Fungus (Alternaria): Morphological and Physiological Characterization. Folia Microbiol. 2010, 55, 393–400. [Google Scholar] [CrossRef]
- Sinharoy, S.; Liu, C.; Breakspear, A.; Guan, D.; Shailes, S.; Nakashima, J.; Zhang, S.; Wen, J.; Torres-Jerez, I.; Oldroyd, G.; et al. A Medicago truncatula Cystathionine-β-Synthase-like Domain-Containing Protein Is Required for Rhizobial Infection and Symbiotic Nitrogen Fixation. Plant Physiol. 2016, 170, 2204–2217. [Google Scholar] [CrossRef]
- Zhang, Y.; Gu, Z.; Ren, Y.; Wang, L.; Zhang, J.; Liang, C.; Tong, S.; Wang, Y.; Xu, D.; Zhang, X.; et al. Integrating Transcriptomics and Metabolomics to Characterize Metabolic Regulation to Elevated CO2 in Chlamydomonas reinhardtii. Mar. Biotechnol. 2021, 23, 255–275. [Google Scholar] [CrossRef]
- Chaiboonchoe, A.; Dohai, B.S.; Cai, H.; Nelson, D.R.; Jijakli, K.; Salehi-Ashtiani, K. Microalgal Metabolic Network Model Refinement through High-Throughput Functional Metabolic Profiling. Front. Bioeng. Biotechnol. 2014, 2, 68. [Google Scholar] [CrossRef]
- Vallon, O.; Bulté, L.; Kuras, R.; Olive, J.; Wollman, F.-A. Extensive Accumulation of an Extracellular L-Amino-Acid Oxidase during Gametogenesis of Chlamydomonas reinhardtii. Eur. J. Biochem. 1993, 215, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Kirk, D.L.; Kirk, M.M. Carrier-Mediated Uptake of Arginine and Urea by Chlamydomonas reinhardtii. Plant Physiol. 1978, 61, 556–560. [Google Scholar] [CrossRef]
- Chaiboonchoe, A.; Ghamsari, L.; Dohai, B.; Ng, P.; Khraiwesh, B.; Jaiswal, A.; Jijakli, K.; Koussa, J.; Nelson, D.R.; Cai, H.; et al. Systems Level Analysis of the Chlamydomonas reinhardtii Metabolic Network Reveals Variability in Evolutionary Co-Conservation. Mol. BioSyst. 2016, 12, 2394–2407. [Google Scholar] [CrossRef] [PubMed]
- Calatrava, V.; Hom, E.F.Y.; Llamas, Á.; Fernández, E.; Galván, A. OK, Thanks! A New Mutualism between Chlamydomonas and Methylobacteria Facilitates Growth on Amino Acids and Peptides. FEMS Microbiol. Lett. 2018, 365, fny021. [Google Scholar] [CrossRef] [PubMed]
- Voigt, J. Biosynthesis and Turnover of Cell Wall Glycoproteins during the Vegetative Cell Cycle of Chlamydomonas reinhardtii. Z. Nat. 1986, 41, 885–896. [Google Scholar]
- Gonzalez-Ballester, D.; Camargo, A.; Fernandez, E. Ammonium Transporter Genes in Chlamydomonas: The Nitrate-Specific Regulatory Gene Nit2 Is Involved in Amt1;1 Expression. Plant Mol. Biol. 2004, 56, 863–878. [Google Scholar] [CrossRef]
- Kim, J.-G.; Park, S.-J.; Sinninghe Damsté, J.S.; Schouten, S.; Irene, W.; Rijpstra, C.; Jung, M.-Y.; Kim, S.-J.; Gwak, J.-H.; Hong, H.; et al. Hydrogen Peroxide Detoxification Is a Key Mechanism for Growth of Ammonia-Oxidizing Archaea. Proc. Natl. Acad. Sci. USA 2016, 113, 7888–7893. [Google Scholar] [CrossRef]
- Hünken, M.; Harder, J.; Kirst, G.O. Epiphytic Bacteria on the Antarctic Ice Diatom Amphiprora kufferathii Manguin Cleave Hydrogen Peroxide Produced during Algal Photosynthesis. Plant Biol. 2008, 10, 519–526. [Google Scholar] [CrossRef]
- Zinser, E.R. Cross-Protection from Hydrogen Peroxide by Helper Microbes: The Impacts on the Cyanobacterium Prochlorococcus and Other Beneficiaries in Marine Communities. Environ. Microbiol. Rep. 2018, 10, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Jamet, A.; Sigaud, S.; Van De Sype, G.; Puppo, A.; Hérouart, D. Expression of the Bacterial Catalase Genes During Sinorhizobium meliloti-Medicago sativa Symbiosis and Their Crucial Role During the Infection Process. Mol. Plant-Microbe Interact. 2003, 16, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Visick, K.L.; Ruby, E.G. The Periplasmic, Group III Catalase of Vibrio fischeri Is Required for Normal Symbiotic Competence and Is Induced Both by Oxidative Stress and by Approach to Stationary Phase. J. Bacteriol. 1998, 180, 2087–2092. [Google Scholar] [CrossRef] [PubMed]
- Keswani, C.; Singh, S.P.; Cueto, L.; García-Estrada, C.; Mezaache-Aichour, S.; Glare, T.R.; Borriss, R.; Singh, S.P.; Angel Blázquez, M.; Sansinenea, E. Auxins of Microbial Origin and Their Use in Agriculture. Appl. Microbiol. Biotechnol. 2020, 104, 8549–8565. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.A.; Hmelo, L.R.; van Tol, H.M.; Durham, B.P.; Carlson, L.T.; Heal, K.R.; Morales, R.L.; Berthiaume, C.T.; Parker, M.S.; Djunaedi, B.; et al. Interaction and Signalling between a Cosmopolitan Phytoplankton and Associated Bacteria. Nature 2015, 522, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Palombella, A.L.; Dutcher, S.K. Identification of the Gene Encoding the Tryptophan Synthase-Subunit from Chlamydomonas Reinhardtii. Plant Physiol. 1998, 117, 455–464. [Google Scholar] [CrossRef]
- De-Bashan, L.E.; Antoun, H.; Bashan, Y. Involvement of Indole-3-Acetic Acid Produced by the Growth-Promoting Bacterium Azospirillum spp. In Promoting Growth of Chlorella vulgaris. J. Phycol. 2008, 44, 938–947. [Google Scholar] [CrossRef]
- Segev, E.; Wyche, T.P.; Kim, K.H.; Petersen, J.; Ellebrandt, C.; Vlamakis, H.; Barteneva, N.; Paulson, J.N.; Chai, L.; Clardy, J.; et al. Dynamic Metabolic Exchange Governs a Marine Algal-Bacterial Interaction. eLife 2016, 5, e17473. [Google Scholar] [CrossRef]
- Palacios, O.A.; Gomez-Anduro, G.; Bashan, Y.; de-Bashan, L.E. Tryptophan, Thiamine and Indole-3-Acetic Acid Exchange between Chlorella sorokiniana and the Plant Growth-Promoting Bacterium Azospirillum brasilense. FEMS Microbiol. Ecol. 2016, 92, fiw077. [Google Scholar] [CrossRef] [PubMed]
- Labeeuw, L.; Khey, J.; Bramucci, A.R.; Atwal, H.; de la Mata, A.P.; Harynuk, J.; Case, R.J. Indole-3-Acetic Acid Is Produced by Emiliania huxleyi Coccolith-Bearing Cells and Triggers a Physiological Response in Bald Cells. Front. Microbiol. 2016, 7, 828. [Google Scholar] [CrossRef]
- Calatrava, V.; Hom, E.F.Y.; Llamas, A.; Fernández, E.; Galván, A. Auxin Production in the Green Alga Chlamydomonas involves an Extracellular L-Amino Acid Oxidase and Supports Algal-Bacterial Mutualism with Methylobacteria. bioRxiv 2022. [Google Scholar] [CrossRef]
- Miller, M.B.; Bassler, B.L. Quorum Sensing in Bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef]
- Li, Q.; Ren, Y.; Fu, X. Inter-Kingdom Signaling between Gut Microbiota and Their Host. Cell. Mol. Life Sci. 2019, 76, 2383–2389. [Google Scholar] [CrossRef]
- Fan, Q.; Wang, H.; Mao, C.; Li, J.; Zhang, X.; Grenier, D.; Yi, L.; Wang, Y. Structure and Signal Regulation Mechanism of Interspecies and Interkingdom Quorum Sensing System Receptors. J. Agric. Food Chem. 2022, 70, 429–445. [Google Scholar] [CrossRef]
- Gahan, C.G.; Patel, S.J.; Chen, L.M.; Manson, D.E.; Ehmer, Z.J.; Blackwell, H.E.; Van Lehn, R.C.; Lynn, D.M. Bacterial Quorum Sensing Signals Promote Large-Scale Remodeling of Lipid Membranes. Langmuir 2021, 37, 9120–9136. [Google Scholar] [CrossRef] [PubMed]
- Givskov, M.; De Nys, R.; Manefield, M.; Gram, L.; Maximilien, R.; Eberl, L.; Molin, S.; Steinberg, P.D.; Kjelleberg, S. Eukaryotic Interference with Homoserine Lactone-Mediated Prokaryotic Signalling. J. Bacteriol. 1996, 178, 6618–6622. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Teplitski, M.; Robinson, J.B.; Bauer, W.D. Production of Substances by Medicago truncatula That Affect Bacterial Quorum Sensing. Mol. Plant-Microbe Interact. 2003, 16, 827–834. [Google Scholar] [CrossRef]
- Teplitski, M.; Chen, H.; Rajamani, S.; Gao, M.; Merighi, M.; Sayre, R.T.; Robinson, J.B.; Rolfe, B.G.; Bauer, W.D. Chlamydomonas reinhardtii Secretes Compounds That Mimic Bacterial Signals and Interfere with Quorum Sensing Regulation in Bacteria. Plant Physiol. 2004, 134, 137–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selim, K.A.; Ermilova, E.; Forchhammer, K. From Cyanobacteria to Archaeplastida: New Evolutionary Insights into PII Signalling in the Plant Kingdom. New Phytol. 2020, 227, 722–731. [Google Scholar] [CrossRef]
- Arcondeguy, T.; Huez, I.; Tillard, P.; Gangneux, C.; de Billy, F.; Gojon, A.; Truchet, G.; Kahn, D. The Rhizobium Meliloti PII Protein, Which Controls Bacterial Nitrogen Metabolism, Affects Alfalfa Nodule Development. Genes Dev. 1997, 11, 1194–1206. [Google Scholar] [CrossRef]
- Rajamani, S.; Bauer, W.D.; Robinson, J.B.; Farrow, J.M.; Pesci, E.C.; Teplitski, M.; Gao, M.; Sayre, R.T.; Phillips, D.A. The Vitamin Riboflavin and Its Derivative Lumichrome Activate the LasR Bacterial Quorum-Sensing Receptor. Mol. Plant-Microbe Interact. 2008, 21, 1184–1192. [Google Scholar] [CrossRef] [PubMed]
- Saijo, Y.; Kossmann, J.; Dakora, F.D.; Matiru, V.N.; Kanu, A.S. Rhizosphere Ecology of Lumichrome and Riboflavin, Two Bacterial Signal Molecules Eliciting Developmental Changes in Plants. Front. Plant Sci. 2015, 6, 700. [Google Scholar] [CrossRef]
- Fabris, M.; Abbriano, R.M.; Pernice, M.; Sutherland, D.L.; Commault, A.S.; Hall, C.C.; Labeeuw, L.; McCauley, J.I.; Kuzhiuparambil, U.; Ray, P.; et al. Emerging Technologies in Algal Biotechnology: Toward the Establishment of a Sustainable, Algae-Based Bioeconomy. Front. Plant Sci. 2020, 11, 279. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, W.; Guo, Y.; Zhang, Z.; Shi, W.; Cui, F.; Lens, P.N.L.; Tay, J.H. Microalgal-Bacterial Consortia: From Interspecies Interactions to Biotechnological Applications. Renew. Sustain. Energy Rev. 2020, 118, 109563. [Google Scholar] [CrossRef]
- Panepinto, D.; Riggio, V.A.; Zanetti, M.; Musmarra, D. Analysis of the Emergent Climate Change Mitigation Technologies. Int. J. Environ. Res. Public Health 2021, 18, 6767. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Baek, J.S.; Yun, Y.S.; Jun Sim, S.; Park, S.; Kim, S.C. Hydrogen Production from Chlamydomonas reinhardtii Biomass Using a Two-Step Conversion Process: Anaerobic Conversion and Photosynthetic Fermentation. Int. J. Hydrog. Energy 2006, 31, 812–816. [Google Scholar] [CrossRef]
- Melis, A.; Melnicki, M.R. Integrated Biological Hydrogen Production. Int. J. Hydrog. Energy 2006, 31, 1563–1573. [Google Scholar] [CrossRef]
- Miyamoto, K.; Ohta, S.; Nawa, Y.; Mori, Y.; Miura, Y. Hydrogen Production by a Mixed Culture of a Green Alga, Chlamydomonas reinhardtii and a Photosynthetic Bacterium, Rhodospirillum rubrum. Agric. Biol. Chem. 1987, 51, 1319–1324. [Google Scholar] [CrossRef]
- Fakhimi, N.; Gonzalez-Ballester, D.; Fernández, E.; Galván, A.; Dubini, A. Algae-Bacteria Consortia as a Strategy to Enhance H2 Production. Cells 2020, 9, 1353. [Google Scholar] [CrossRef]
- Fakhimi, N.; Dubini, A.; Tavakoli, O.; González-Ballester, D. Acetic Acid Is Key for Synergetic Hydrogen Production in Chlamydomonas-Bacteria Co-Cultures. Bioresour. Technol. 2019, 289, 121648. [Google Scholar] [CrossRef]
- Fakhimi, N.; Tavakoli, O.; Marashi, S.A.; Moghimi, H.; Mehrnia, M.R.; Dubini, A.; González-Ballester, D. Acetic Acid Uptake Rate Controls H2 Production in Chlamydomonas-Bacteria Co-Cultures. Algal Res. 2019, 42, 101605. [Google Scholar] [CrossRef]
- Edrei, J. Methods of Generating Hydrogen. Application US13/582,442, 27 December 2012. Available online: http://www.freepatentsonline.com/y2012/0329089.html (accessed on 27 December 2012).
- Xu, L.; Cheng, X.; Wang, Q. Effect of Co-Cultivation of Chlamydomonas Reinhardtii with Azotobacter chroococcum on Hydrogen Production. Int. J. Hydrog. Energy 2017, 42, 22713–22719. [Google Scholar] [CrossRef]
- Yu, Q.; He, J.; Zhao, Q.; Wang, X.; Zhi, Y.; Li, X.; Li, X.; Li, L.; Ge, B. Regulation of Nitrogen Source for Enhanced Photobiological H2 Production by Co-Culture of Chlamydomonas reinhardtii and Mesorhizobium sangaii. Algal Res. 2021, 58, 102422. [Google Scholar] [CrossRef]
- Wu, S.; Li, X.; Yu, J.; Wang, Q. Increased Hydrogen Production in Co-Culture of Chlamydomonas reinhardtii and Bradyrhizobium japonicum. Bioresour. Technol. 2012, 123, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.J.; Gonzalez-Ballester, D.; Gomez-Osuna, A.; Galván, A.; Fernandez, E.; Dubini, A. Chlamydomonas-Methylobacterium oryzae Cooperation Leads to Increased Biomass, Nitrogen Removal, and Hydrogen Production. Bioresour. Technol. 2022, 352, 127088. [Google Scholar] [CrossRef]
- Lakatos, G.; Deák, Z.; Vass, I.; Retfalvi, T.; Rozgonyi, S.; Rákhely, G.; Ördög, V.; Kondorosi, E.; Maróti, G. Bacterial Symbionts Enhance Photo-Fermentative Hydrogen Evolution of Chlamydomonas Algae. Green Chem. 2014, 16, 4716–4727. [Google Scholar] [CrossRef]
- Xu, L.; Li, D.; Wang, Q.; Wu, S. Improved Hydrogen Production and Biomass through the Co-Cultivation of Chlamydomonas reinhardtii and Bradyrhizobium japonicum. Int. J. Hydrog. Energy 2016, 41, 9276–9283. [Google Scholar] [CrossRef]
- Pachapur, V.L.; Sarma, S.J.; Brar, S.K.; Bihan, Y.L.; Soccol, C.R.; Buelna, G.; Verma, M. Co-Culture Strategies for Increased Biohydrogen Production. Int. J. Energy Res. 2015, 39, 1479–1504. [Google Scholar] [CrossRef]
- Sivaramakrishnan, R.; Suresh, S.; Kanwal, S.; Ramadoss, G.; Ramprakash, B.; Incharoensakdi, A. Microalgal Biorefinery Concepts’ Developments for Biofuel and Bioproducts: Current Perspective and Bottlenecks. Int. J. Mol. Sci. 2022, 23, 2623. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Li, T.; Wang, Q. Ten Years of Algal Biofuel and Bioproducts: Gains and Pains. Planta 2019, 249, 195–219. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, J.L.; Garbayo, I.; Cuaresma, M.; Montero, Z.; González-Del-Valle, M.; Vílchez, C. Impact of Microalgae-Bacteria Interactions on the Production of Algal Biomass and Associated Compounds. Mar. Drugs 2016, 14, 100. [Google Scholar] [CrossRef]
- Xu, L.; Cheng, X.; Wang, Q. Enhanced Lipid Production in Chlamydomonas reinhardtii by Co-Culturing With Azotobacter chroococcum. Front. Plant Sci. 2018, 9, 741. [Google Scholar] [CrossRef]
- Holland, M.; Di Bello, P.; Carlton, R.M. Method for Increasing Algae Growth and the Use Thereof in Production of Algae-Derived Biofuels and Other Chemical. U.S. Patent No. 8,778,660 B2, 15 July 2014. [Google Scholar]
- Zuccaro, G.; Steyer, J.P.; van Lis, R. The Algal Trophic Mode Affects the Interaction and Oil Production of a Synergistic Microalga-Yeast Consortium. Bioresour. Technol. 2019, 273, 608–617. [Google Scholar] [CrossRef]
- Nishio, K.; Hashimoto, K.; Watanabe, K. Digestion of Algal Biomass for Electricity Generation in Microbial Fuel Cells. Biosci. Biotechnol. Biochem 2013, 3, 670–672. [Google Scholar] [CrossRef]
- Bélanger-Lépine, F.; Tremblay, A.; Huot, Y.; Barnabé, S. Cultivation of an Algae-Bacteria Consortium in Wastewater from an Industrial Park: Effect of Environmental Stress and Nutrient Deficiency on Lipid Production. Bioresour. Technol. 2018, 267, 657–665. [Google Scholar] [CrossRef]
- Mantovani, M.; Marazzi, F.; Fornaroli, R.; Bellucci, M.; Ficara, E.; Mezzanotte, V. Outdoor Pilot-Scale Raceway as a Microalgae-Bacteria Sidestream Treatment in a WWTP. Sci. Total Environ. 2020, 710, 135583. [Google Scholar] [CrossRef] [PubMed]
- Oruganti, R.K.; Katam, K.; Show, P.L.; Gadhamshetty, V.; Upadhyayula, V.K.K.; Bhattacharyya, D. A Comprehensive Review on the Use of Algal-Bacterial Systems for Wastewater Treatment with Emphasis on Nutrient and Micropollutant Removal. Bioengineered 2022, 13, 10412–10453. [Google Scholar] [CrossRef] [PubMed]
- Viswanaathan, S.; Perumal, P.K.; Sundaram, S. Integrated Approach for Carbon Sequestration and Wastewater Treatment Using Algal–Bacterial Consortia: Opportunities and Challenges. Sustainability 2022, 14, 1075. [Google Scholar] [CrossRef]
- Toyama, T.; Kasuya, M.; Hanaoka, T.; Kobayashi, N.; Tanaka, Y.; Inoue, D.; Sei, K.; Morikawa, M.; Mori, K. Growth Promotion of Three Microalgae, Chlamydomonas reinhardtii, Chlorella vulgaris and Euglena gracilis, by in Situ Indigenous Bacteria in Wastewater Effluent. Biotechnol. Biofuels 2018, 11, 176. [Google Scholar] [CrossRef]
- Jia, H.; Yuan, Q. Removal of Nitrogen from Wastewater Using Microalgae and Microalgae–Bacteria Consortia. Cogent Environ. Sci. 2016, 2, 1275089. [Google Scholar] [CrossRef]
- Ashok, V.; Shriwastav, A.; Bose, P. Nutrient Removal Using Algal-Bacterial Mixed Culture. Appl. Biochem. Biotechnol. 2014, 174, 2827–2838. [Google Scholar] [CrossRef]
- Mironiuk, M.; Chojnacka, K.W. The Environmental Benefits Arising from the Use of Algae Biomass in Industry. Algae Biomass Charact. Appl. 2018, 8, 7–16. [Google Scholar]
- Wangpraseurt, D.; You, S.; Sun, Y.; Chen, S. Biomimetic 3D Living Materials Powered by Microorganisms. Trends Biotechnol. 2022, 40, 843–857. [Google Scholar] [CrossRef] [PubMed]
- Das, A.A.K.; Bovill, J.; Ayesh, M.; Stoyanov Bc, S.D.; Paunov, V.N. Fabrication of Living Soft Matter by Symbiotic Growth of Unicellular Microorganisms. J. Mater. Chem. B 2016, 4, 3685–3694. [Google Scholar] [CrossRef]
- D’Adamo, S.; Kormelink, R.; Martens, D.; Barbosa, M.J.; Wijffels, R.H. Prospects for Viruses Infecting Eukaryotic Microalgae in Biotechnology. Biotechnol. Adv. 2022, 54, 107790. [Google Scholar] [CrossRef] [PubMed]
- Alcántara-Martínez, N.; Figueroa-Martínez, F.; Rivera-Cabrera, F.; Volke-Sepúlveda, T. An Unexpected Guest: A Green Microalga Associated with the Arsenic-Tolerant Shrub Acacia farnesiana. FEMS Microbiol. Ecol. 2022, 98, fiac041. [Google Scholar] [CrossRef] [PubMed]
- Cirri, E.; Pohnert, G. Algae−bacteria Interactions That Balance the Planktonic Microbiome. New Phytologist 2019, 223, 100–106. [Google Scholar] [CrossRef] [Green Version]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calatrava, V.; Tejada-Jimenez, M.; Sanz-Luque, E.; Fernandez, E.; Galvan, A.; Llamas, A. Chlamydomonas reinhardtii, a Reference Organism to Study Algal–Microbial Interactions: Why Can’t They Be Friends? Plants 2023, 12, 788. https://doi.org/10.3390/plants12040788
Calatrava V, Tejada-Jimenez M, Sanz-Luque E, Fernandez E, Galvan A, Llamas A. Chlamydomonas reinhardtii, a Reference Organism to Study Algal–Microbial Interactions: Why Can’t They Be Friends? Plants. 2023; 12(4):788. https://doi.org/10.3390/plants12040788
Chicago/Turabian StyleCalatrava, Victoria, Manuel Tejada-Jimenez, Emanuel Sanz-Luque, Emilio Fernandez, Aurora Galvan, and Angel Llamas. 2023. "Chlamydomonas reinhardtii, a Reference Organism to Study Algal–Microbial Interactions: Why Can’t They Be Friends?" Plants 12, no. 4: 788. https://doi.org/10.3390/plants12040788
APA StyleCalatrava, V., Tejada-Jimenez, M., Sanz-Luque, E., Fernandez, E., Galvan, A., & Llamas, A. (2023). Chlamydomonas reinhardtii, a Reference Organism to Study Algal–Microbial Interactions: Why Can’t They Be Friends? Plants, 12(4), 788. https://doi.org/10.3390/plants12040788