SERS of Human Red Blood Cells in Non-Resonant Conditions: Benefits, Limitations, and Complementary Tools (CytoViva and GFAAS)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Synthesis of Silver Nanoparticles (AgNPs)
2.3. Tangential Flow Filtration (TFF) of AgNPs
2.4. Exposure of Human Red Blood Cells (RBCs) to AgNPs, Cell Counting, and Hematocrit
2.5. Transmission Electron Microscopy (TEM) of AgNPs
2.6. UV-Vis Absorption Spectrophotometry of AgNPs and RBCs
2.7. Raman and Surface-Enhanced Raman Spectroscopy (SERS) of AgNPs and RBCs
2.8. CytoViva Hyperspectral Imaging of AgNPs and RBCs
2.9. Inductively Coupled Plasma–Optical Emission Spectroscopy (ICP-OES) and Graphite Furnace Atomic Absorption Spectroscopy (GFAAS) of AgNPs and of RBCs
2.10. Statistics
3. Results
3.1. Characterization of AgNPs
3.2. RBC Absorption Spectra, Counting, and Hematocrit
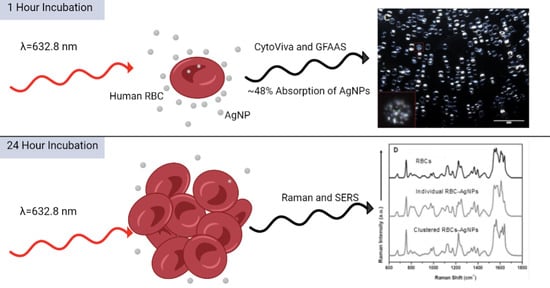
3.3. Raman and Surface-Enhanced Raman Spectroscopy (SERS) of AgNPs and RBCs
3.4. CytoViva Hyperspectral Imaging of AgNPs and RBCs
3.5. ICP-OES and GFAAS of AgNPs and RBCs
4. Discussion
4.1. Raman and Surface-Enhanced Raman Spectroscopy (SERS) of AgNPs and RBCs
4.2. CytoViva Hyperspectral Imaging of AgNPs and RBCs
4.3. ICP-OES and GFAAS of AgNPs and RBCs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qiu, Y.; Kuang, C.; Liu, X.; Tang, L. Single-molecule surface-enhanced Raman spectroscopy. Sensors 2022, 22, 4889. [Google Scholar] [CrossRef] [PubMed]
- Moskovits, M. Surface-enhanced Raman spectroscopy: A brief retrospective. J. Raman Spectrosc. 2005, 36, 485–496. [Google Scholar] [CrossRef]
- Kneipp, K. Surface-enhanced Raman scattering. Phys. Today 2007, 60, 40–46. [Google Scholar] [CrossRef] [Green Version]
- Blackie, E.J.; Le Ru, E.C.; Etchegoin, P.G. Single-molecule surface-enhanced Raman spectroscopy of nonresonant molecules. J. Am. Chem. Soc. 2009, 131, 14466–14472. [Google Scholar] [CrossRef]
- Xu, H.; Aizpurua, J.; Käll, M.; Apell, P. Electromagnetic contributions to single-molecule sensitivity in surface-enhanced Raman spectroscopy. Phys. Rev. E 2000, 62, 4318–4324. [Google Scholar] [CrossRef] [Green Version]
- Braun, G.; Pavel, I.; Morrill, A.R.; Seferos, D.S.; Bazan, G.C.; Reich, N.O.; Moskovits, M. Chemically patterned microspheres for controlled nanoparticle assembly in the construction of SERS hot spots. J. Am. Chem. Soc. 2007, 129, 7760–7761. [Google Scholar] [CrossRef]
- Kim, N.; Hwang, W.; Baek, K.; Rohman, M.R.; Kim, J.; Kim, H.W.; Mun, J.; Lee, S.Y.; Yun, G.; Murray, J.; et al. Smart SERS hot spots: Single molecules can be positioned in a plasmonic nanojunction using host-guest chemistry. J. Am. Chem. Soc. 2018, 140, 4705–4711. [Google Scholar] [CrossRef]
- Tan, Y.; Yan, B.; Xue, L.; Li, Y.; Luo, X.; Ji, P. Surface-enhanced Raman spectroscopy of blood serum based on gold nanoparticles for the diagnosis of the oral squamous cell carcinoma. Lipids Health Dis. 2017, 16, 73. [Google Scholar] [CrossRef] [Green Version]
- Boyd, S.; Massimo, B.F.; Bertino, F.; Ye, D.; White, L.S.; Seashols, S.J. Highly sensitive detection of blood by surface enhanced Raman scattering. J. Forensic. Sci. 2013, 58, 753–756. [Google Scholar] [CrossRef]
- Wood, B.R.; Kochan, K.; Marzec, K.M. Resonance Raman spectroscopy of hemoglobin in red blood cells. In VibrationalSpectroscopy in Protein Research, 1st ed.; Ozaki, Y., Baranska, M., Lednev, I.K., Wood, B.R., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2020; pp. 375–414. [Google Scholar]
- Dybas, J.; Alcicek, F.C.; Wajda, A.; Kaczmarska, M.; Zimna, A.; Bulat, K.; Blat, A.; Stepanenko, T.; Mohaissen, T.; Szczesny-Malysiak, E.; et al. Trends in biomedical analysis of red blood cells—Raman spectroscopy against other spectroscopic, microscopic and classical techniques. TrAC 2022, 146, 116481. [Google Scholar] [CrossRef]
- Barkur, S.; Chidangil, S. Surface-enhanced Raman spectroscopy study of red blood cells and platelets. J. Biomol. Struct. Dyn. 2019, 37, 1090–1098. [Google Scholar] [CrossRef] [PubMed]
- Harada, I.; Takeuchi, H. Raman and ultraviolet resonance Raman spectra of proteins and related compounds. In Spectroscopy of Biological Systems, 1st ed.; Clark, R.J.H., Hester, R.E., Eds.; John Wiley & Sons: Chichester, UK, 1986; Volume 13, pp. 113–175. [Google Scholar]
- Drescher, D.; Büchner, T.; McNaughton, D.; Kneipp, J. SERS reveals the specific interaction of silver and gold nanoparticles with hemoglobin and red blood cell components. Phys. Chem. Chem. Phys. 2013, 15, 5364–5373. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Smith, K.M.; Spiro, T.G. Assignment of protoheme resonance Raman spectrum by heme labeling in myoglobin. J. Am. Chem. Soc. 1996, 118, 12638–12646. [Google Scholar] [CrossRef]
- Wood, B.R.; Caspers, P.; Puppels, G.J.; Pandiancherri, S.; McNaughton, D. Resonance Raman spectroscopy of red blood cells using near-infrared laser excitation. Anal. Bioanal. Chem. 2007, 387, 1691–1703. [Google Scholar] [CrossRef] [PubMed]
- Bankapur, A.; Barkur, S.; Chidangil, S.; Mathur, D. A micro-Raman study of live, single red blood cells (RBCs) treated with AgNO3 nanoparticles. PLoS ONE 2014, 9, e103493. [Google Scholar] [CrossRef] [Green Version]
- Premasiri, W.R.; Lee, J.C.; Ziegler, L.D. Surface-enhanced Raman scattering of whole human blood, blood plasma, and red blood cells: Cellular processes and bioanalytical sensing. J. Phys. Chem. B 2012, 116, 9376–9386. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Flaherty, B.R.; Cohen, C.E.; Peterson, D.S.; Zhao, Y. Direct detection of malaria-infected red blood cells by surface-enhanced Raman spectroscopy. Nanomedicine 2016, 12, 1445–1451. [Google Scholar] [CrossRef] [Green Version]
- Lee, P.C.; Meisel, D. Adsorption and surface-enhanced Raman of dyes on silver and gold sols. J. Phys. Chem. 1982, 86, 3391–3395. [Google Scholar] [CrossRef]
- Wan, Y.; Guo, Z.; Jiang, X.; Fang, K.; Lu, X.; Zhang, Y.; Gu, N. Quasi-spherical silver nanoparticles: Aqueous synthesis and size control by the seed-mediated Lee-Meisel method. J. Colloid Interface Sci. 2013, 394, 263–268. [Google Scholar] [CrossRef]
- Trefry, J.C.; Monahan, J.L.; Weaver, K.M.; Meyerhoefer, A.J.; Markopolous, M.M.; Arnold, Z.S.; Wooley, D.P.; Pavel, I.E. Size selection and concentration of silver nanoparticles by tangential flow ultrafiltration for SERS-based biosensors. J. Am. Chem. Soc. 2010, 132, 10970–10972. [Google Scholar] [CrossRef]
- Anders, C.B.; Baker, J.D.; Stahler, A.C.; Williams, A.J.; Sisco, J.N.; Trefry, J.C.; Wooley, D.P.; Sizemore, I.E. Tangential flow ultrafiltration: A “green” method for the size selection and concentration of colloidal silver nanoparticles. J. Vis. Exp. 2012, 68, e4167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, C.; Lumb, A.B. Physiology of haemoglobin. CEACCP 2012, 12, 251–256. [Google Scholar] [CrossRef]
- Gell, D.A. Structure and function of haemoglobines. Blood Cells Mol. Dis. 2018, 70, 13–42. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.H.; Ghatge, M.S.; Safo, M.K. Hemoglobin: Structure, function, and allostery. Subcell Biochem. 2020, 94, 345–382. [Google Scholar] [PubMed]
- Israelsen, N.D.; Hanson, C.; Vargis, E. Nanoparticle properties and synthesis effects on surface-enhanced Raman scattering enhancement factor: An introduction. Sci. World J. 2015, 2015, 124582. [Google Scholar] [CrossRef] [Green Version]
- Dorney, K. A Chemical-Free Approach for Increasing the Biochemical Surface-Enhanced Raman Spectroscopy (SERS)-Based Sensing Capabilities of Colloidal Silver Nanoparticles. Master’s Thesis, Wright State University, Dayton, OH, USA, 2014. [Google Scholar]
- Stamplecoskie, K.G.; Scaiano, J.C.; Tiwari, V.S.; Anis, H. Optimal size of silver nanoparticles for surface-enhanced Raman spectroscopy. J. Phys. Chem. C 2011, 115, 1403–1409. [Google Scholar] [CrossRef]
- Chen, L.Q.; Fang, L.; Ling, J.; Ding, C.Z.; Kang, B.; Huang, C.Z. Nanotoxicity of silver nanoparticles to red blood cells: Size dependent adsorption, uptake, and hemolytic activity. Chem. Res. Toxicol. 2015, 28, 501–509. [Google Scholar] [CrossRef]
- Bhat, A.; Huan, K.; Cooks, T.; Boukari, H.; Lu, Q. Probing interactions between AuNPs/AgNPs and giant unilamellar vesicles (GUVs) using hyperspectral dark-field microscopy. Int. J. Mol. Sci. 2018, 19, 1014. [Google Scholar] [CrossRef] [Green Version]
- Kettler, K.; Krystek, P.; Giannakou, C.; Hendricks, A.J.; de Jong, W.H. Exploring the effect of silver nanoparticle size and medium composition on uptake into pulmonary epithelial 16HBE14o-cells. J. Nanopart. Res. 2016, 18, 182. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Bai, J.; Jiang, X.; Nienhaus, G.U. Cellular Uptake of Nanoparticles by Membrane Penetration: A Study Combining Confocal Microscopy with FTIR Spectroelectrochemistry. ACS Nano 2012, 6, 1251–1259. [Google Scholar] [CrossRef]
- Geiser, M.; Rothen-Rutishauser, B.; Kapp, N.; Schuerch, S.; Kreyling, W.; Schulz, H.; Semmler, M.; Im Hof, V.; Heyder, J.; Gehr, P. Ultrafine particles cross cellular membranes by nonphagocytic mechanisms in lungs and in cultured cells. Environ. Health Perspect. 2005, 113, 1555–1560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leroueil, P.R.; Berry, S.A.; Duthie, K.; Han, G.; Rotello, V.M.; McNerny, D.Q.; Bakerm, J.R., Jr.; Orr, B.G.; Holl, M.M. Wide varieties of cationic nanoparticles induce defects in supported lipid bilayers. Nano Lett. 2008, 8, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Giridhar, G.; Manepalli, R.R.K.N.; Apparao, G. Chapter 7—Confocal Raman Spectroscopy. In Spectroscopic Methods for Nanomaterials Characterization; Elsevier: Amsterdam, The Netherlands, 2017; pp. 141–161. [Google Scholar] [CrossRef]
- Elliot, A.D. Confocal Microscopy: Principles and Modern Practices. Curr. Protoc. Cytom. 2020, 92, e68. [Google Scholar] [CrossRef]
- CytoViva in CytoViva Hyperspectral Microscope at CytoViva | Enhanced Darkfield Hyperspectral Microscope | Products. Available online: https://www.cytoviva.com (accessed on 18 June 2023).
- Aguilar, Z.P. Nanomaterials for Medical Applications; Elsevier: Amsterdam, The Netherlands, 2013; 2.1.9: Atomic Absorption Spectroscopy. [Google Scholar] [CrossRef]
- Svitkova, B.; Selc, M.; Nemethova, V.; Razga, F.; Gabelova, A.; Ursinyova, M.; Babelova, A. Plate reader spectroscopy as an alternative to atomic absorption spectroscopy for the assessment of nanoparticles cellular uptake. Heliyon 2022, 8, e11595. [Google Scholar] [CrossRef]
- Flores, C.Y.; Minan, A.G.; Grillo, C.A.; Salvarezza, R.C.; Vericat, C.; Schilardi, P.L. Citrate-capped silver nanoparticles showing good bacterial effect against both plankytonic and sessile bacteria and a low cytotoxicity to osteoblastic cells. ACS Appl. Mater. Interfaces 2013, 5, 3149–3159. [Google Scholar] [CrossRef]
- Ma, C.; Gerhard, E.; Lu, D.; Yang, J. Citrate chemistry and biology for biomaterials design. Biomaterials 2018, 178, 383–400. [Google Scholar] [CrossRef] [PubMed]





| “Blue” AgNPs | Band Position (nm) | FWHM (nm) |
|---|---|---|
| Control | 464 | 60.6 ± 0.5 |
| Intracellular | 479 | 153 ± 2 |
| Extracellular | 490 | 142 ± 1 |
| Sample | Amount of Ag (µg mL−1) | Mass Percentage of AgNPs |
|---|---|---|
| Incubation mixture | 34.00 ± 0.74 | 100% |
| Incubation supernatant | 16.34 ± 2.30 | ~48.1% |
| Post-wash supernatants #1–3 | 0.825 ± 0.768 | ~2.4% |
| RBCs-AgNPs | 16.24 ± 1.39 | ~47.8% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wells, K.L.; Alla, P.K.; Kaiser, K.G.; Murgulet, I.T.; Adragna, N.C.; Pavel, I.E. SERS of Human Red Blood Cells in Non-Resonant Conditions: Benefits, Limitations, and Complementary Tools (CytoViva and GFAAS). Chemosensors 2023, 11, 353. https://doi.org/10.3390/chemosensors11070353
Wells KL, Alla PK, Kaiser KG, Murgulet IT, Adragna NC, Pavel IE. SERS of Human Red Blood Cells in Non-Resonant Conditions: Benefits, Limitations, and Complementary Tools (CytoViva and GFAAS). Chemosensors. 2023; 11(7):353. https://doi.org/10.3390/chemosensors11070353
Chicago/Turabian StyleWells, Kelsey L., Praveen K. Alla, Kyra G. Kaiser, Ioana T. Murgulet, Norma C. Adragna, and Ioana E. Pavel. 2023. "SERS of Human Red Blood Cells in Non-Resonant Conditions: Benefits, Limitations, and Complementary Tools (CytoViva and GFAAS)" Chemosensors 11, no. 7: 353. https://doi.org/10.3390/chemosensors11070353
APA StyleWells, K. L., Alla, P. K., Kaiser, K. G., Murgulet, I. T., Adragna, N. C., & Pavel, I. E. (2023). SERS of Human Red Blood Cells in Non-Resonant Conditions: Benefits, Limitations, and Complementary Tools (CytoViva and GFAAS). Chemosensors, 11(7), 353. https://doi.org/10.3390/chemosensors11070353