Chemical Barrier Proteins in Human Body Fluids
Abstract
:1. Introduction
2. Members of the Chemical Barriers
2.1. Prototypic AMPs in the Chemical Barriers
2.2. Highly Abundant Body Fluid Proteins Are Constituents of the Chemical Barriers
2.3. AMPs with Lower Abundance in the Chemical Barriers
2.4. Proteases and Protease Inhibitors
2.5. Role of AMPs in Nosocomial Infections
3. Chemical Barrier Proteins in Human Body Fluids
3.1. The Composition of the Chemical Barrier in Serum
3.2. Tears, the Chemical Barrier of the Eye
3.3. Salivary Proteins in the Defense of the Oral Cavity
3.4. Sweat–The Chemical Barrier of the Skin
3.5. The Chemical Barrier of the Nasal Secretion
3.6. AMPs Secreted into the Urine
3.7. Antimicrobial and Immunomodulatory Properties of Cervicovaginal Fluid
3.8. Chemical Barrier Proteins in the Seminal Fluid
3.9. AMPs in the Cerebrospinal Fluid
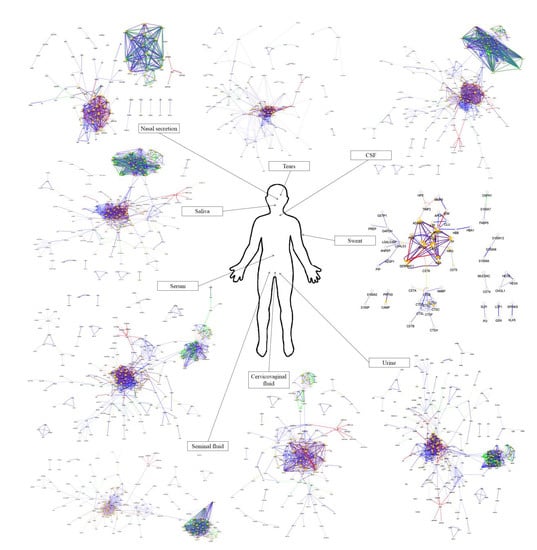
4. Comparison of the Protein–Protein Interaction Network in the Examined Body Fluids
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sperandio, B.; Fischer, N.; Sansonetti, P.J. Mucosal physical and chemical innate barriers: Lessons from microbial evasion strategies. Semin. Immunol. 2015, 27, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Wiesner, J.; Vilcinskas, A. Antimicrobial peptides: The ancient arm of the human immune system. Virulence 2010, 1, 440–464. [Google Scholar] [CrossRef] [PubMed]
- Kalló, G.; Emri, M.; Varga, Z.; Ujhelyi, B.; Tőzsér, J.; Csutak, A.; Csősz, É. Changes in the Chemical Barrier Composition of Tears in Alzheimer’s Disease Reveal Potential Tear Diagnostic Biomarkers. PLoS ONE 2016, 11, e0158000. [Google Scholar] [CrossRef] [Green Version]
- Brandwein, M.; Bentwich, Z.; Steinberg, D. Endogenous Antimicrobial Peptide Expression in Response to Bacterial Epidermal Colonization. Front. Immunol. 2017, 8, 27. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Zhao, S.Z.; Koh, S.K.; Chen, L.; Vaz, C.; Tanavde, V.; Li, X.R.; Beuerman, R.W. In-depth analysis of the human tear proteome. J. Proteom. 2012, 75, 3877–3885. [Google Scholar] [CrossRef]
- Csősz, É.; Emri, G.; Kalló, G.; Tsaprailis, G.; Tőzsér, J. Highly abundant defense proteins in human sweat as revealed by targeted proteomics and label-free quantification mass spectrometry. J. Eur. Acad. Dermatol. Venereol. 2015, 29, 2024–2031. [Google Scholar] [CrossRef] [PubMed]
- Arzumanyan, V.G.; Ozhovan, I.M.; Svitich, O.A. Antimicrobial Effect of Albumin on Bacteria and Yeast Cells. Bull. Exp. Biol. Med. 2019, 167, 763–766. [Google Scholar] [CrossRef]
- Parish, C.A.; Jiang, H.; Tokiwa, Y.; Berova, N.; Nakanishi, K.; McCabe, D.; Zuckerman, W.; Xia, M.M.; Gabay, J.E. Broad-spectrum antimicrobial activity of hemoglobin. Bioorg. Med. Chem. 2001, 9, 377–382. [Google Scholar] [CrossRef]
- Petrlova, J.; Petruk, G.; Huber, R.G.; McBurnie, E.W.; van der Plas, M.J.A.; Bond, P.J.; Puthia, M.; Schmidtchen, A. Thrombin-derived C-terminal fragments aggregate and scavenge bacteria and their proinflammatory products. J. Biol. Chem. 2020, 295, 3417–3430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mantis, N.J.; Rol, N.; Corthésy, B. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011, 4, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Von Deuster, C.I.E.; Knecht, V. Competing interactions for antimicrobial selectivity based on charge complementarity. Biochim. Biophys. Acta-Biomembr. 2011, 1808, 2867–2876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarczak, J.; Kościuczuk, E.M.; Lisowski, P.; Strzałkowska, N.; Jóźwik, A.; Horbańczuk, J.; Krzyzewski, J.; Zwierzchowski, L.; Bagnicka, E. Defensins: Natural component of human innate immunity. Hum. Immunol. 2013, 74, 1069–1079. [Google Scholar] [CrossRef]
- Ganz, T. Defensins: Antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003, 3, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Andreas, B.; Heiner, S. Human alpha-defensin 1 (HNP-1) inhibits adenoviral infection in vitro. Regul. Pept. 2001, 101, 157–161. [Google Scholar]
- Lehrer, R.I.; Ganz, T. Defensins of vertebrate animals. Curr. Opin. Immunol. 2002, 14, 96–102. [Google Scholar] [CrossRef]
- Chen, H.; Xu, Z.; Peng, L.; Fang, X.; Yin, X.; Xu, N.; Cen, P. Recent advances in the research and development of human defensins. Peptides 2006, 27, 931–940. [Google Scholar] [CrossRef]
- Yang, D.; Chertov, O.; Bykovskaia, S.N.; Chen, Q.; Buffo, M.J.; Shogan, J.; Anderson, M.; Schröder, J.M.; Wang, J.M.; Howard, O.M.; et al. Beta-defensins: Linking innate and adaptive immunity through dendritic and T cell CCR6. Science 1999, 286, 525–528. [Google Scholar] [CrossRef]
- Radek, K.; Gallo, R. Antimicrobial peptides: Natural effectors of the innate immune system. Semin. Immunopathol. 2007, 29, 27–43. [Google Scholar] [CrossRef]
- Li, D.; Wang, W.; Shi, H.; Fu, Y.; Chen, X.; Chen, X.; Liu, Y.; Kan, B.; Wang, Y. Gene therapy with beta-defensin 2 induces antitumor immunity and enhances local antitumor effects. Hum. Gene Ther. 2014, 25, 63–72. [Google Scholar] [CrossRef] [Green Version]
- Auvynet, C.; Rosenstein, Y. Multifunctional host defense peptides: Antimicrobial peptides, the small yet big players in innate and adaptive immunity. FEBS J. 2009, 276, 6497–6508. [Google Scholar] [CrossRef]
- De Smet, K.; Contreras, R. Human antimicrobial peptides: Defensins, cathelicidins and histatins. Biotechnol. Lett. 2005, 27, 1337–1347. [Google Scholar] [CrossRef]
- Heilborn, J.D.; Nilsson, M.F.; Kratz, G.; Weber, G.; Sørensen, O.; Borregaard, N.; Ståhle-Bäckdahl, M. The cathelicidin anti-microbial peptide LL-37 is involved in re-epithelialization of human skin wounds and is lacking in chronic ulcer epithelium. J. Investig. Dermatol. 2003, 120, 379–389. [Google Scholar] [CrossRef] [Green Version]
- Yamasaki, K.; Schauber, J.; Coda, A.; Lin, H.; Dorschner, R.A.; Schechter, N.M.; Bonnart, C.; Descargues, P.; Hovnanian, A.; Gallo, R.L. Kallikrein-mediated proteolysis regulates the antimicrobial effects of cathelicidins in skin. FASEB J. 2006, 20, 2068–2080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruni, N.; Capucchio, M.T.; Biasibetti, E.; Pessione, E.; Cirrincione, S.; Giraudo, L.; Corona, A.; Dosio, F. Antimicrobial Activity of Lactoferrin-Related Peptides and Applications in Human and Veterinary Medicine. Molecules 2016, 21, 752. [Google Scholar] [CrossRef]
- Aizawa, S.; Hoki, M.; Yamamuro, Y. Lactoferrin promotes autophagy via AMP-activated protein kinase activation through low-density lipoprotein receptor-related protein 1. Biochem. Biophys. Res. Commun. 2017, 493, 509–513. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Furmanski, P. Sequence specificity and transcriptional activation in the binding of lactoferrin to DNA. Nature 1995, 373, 721–724. [Google Scholar] [CrossRef]
- Shimada, T.; Park, B.G.; Wolf, A.J.; Brikos, C.; Goodridge, H.S.; Becker, C.A.; Reyes, C.N.; Miao, E.A.; Aderem, A.; Götz, F.; et al. Staphylococcus aureus evades lysozyme-based peptidoglycan digestion that links phagocytosis, inflammasome activation, and IL-1beta secretion. Cell Host Microbe 2010, 7, 38–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marquis, G.; Garzon, S.; Strykowski, H.; Auger, P. Cell walls of normal and lysozyme-damaged blastoconidia of Candida albicans: Localization of surface factor 4 antigen and vicinal-glycol staining. Infect. Immun. 1991, 59, 1312–1318. [Google Scholar] [CrossRef] [Green Version]
- Lee-Huang, S.; Huang, P.L.; Sun, Y.; Kung, H.F.; Blithe, D.L.; Chen, H.C. Lysozyme and RNases as anti-HIV components in beta-core preparations of human chorionic gonadotropin. Proc. Natl. Acad. Sci. USA 1999, 96, 2678–2681. [Google Scholar] [CrossRef] [Green Version]
- Tagashira, A.; Nishi, K.; Matsumoto, S.; Sugahara, T. Anti-inflammatory effect of lysozyme from hen egg white on mouse peritoneal macrophages. Cytotechnology 2018, 70, 929. [Google Scholar] [CrossRef]
- Flo, T.H.; Smith, K.D.; Sato, S.; Rodriguez, D.J.; Holmes, M.A.; Strong, R.K.; Akira, S.; Aderem, A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 2004, 432, 917–921. [Google Scholar] [CrossRef]
- Yang, J.; Goetz, D.; Li, J.Y.; Wang, W.; Mori, K.; Setlik, D.; Du, T.; Erdjument-Bromage, H.; Tempst, P.; Strong, R.; et al. An iron delivery pathway mediated by a lipocalin. Mol. Cell 2002, 10, 1045–1056. [Google Scholar] [CrossRef]
- Du, Z.P.; Wu, B.L.; Wu, X.; Lin, X.H.; Qiu, X.Y.; Zhan, X.F.; Wang, S.H.; Shen, J.H.; Zheng, C.P.; Wu, Z.Y.; et al. A systematic analysis of human lipocalin family and its expression in esophageal carcinoma. Sci. Rep. 2015, 5, 12010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bratt, T. Lipocalins and cancer. Biochim. Biophys. Acta 2000, 1482, 318–326. [Google Scholar] [CrossRef]
- Santiago-Sánchez, G.S.; Pita-Grisanti, V.; Quiñones-Díaz, B.; Gumpper, K.; Cruz-Monserrate, Z.; Vivas-Mejía, P.E. Biological Functions and Therapeutic Potential of Lipocalin 2 in Cancer. Int. J. Mol. Sci. 2020, 21, 4365. [Google Scholar] [CrossRef]
- Mesquita, S.D.; Ferreira, A.C.; Falcao, A.M.; Sousa, J.C.; Oliveira, T.G.; Correia-Neves, M.; Sousa, N.; Marques, F.; Palha, J.A. Lipocalin 2 modulates the cellular response to amyloid beta. Cell Death Differ. 2014, 21, 1588–1599. [Google Scholar] [CrossRef] [Green Version]
- McKown, R.L.; Wang, N.; Raab, R.W.; Karnati, R.; Zhang, Y.; Williams, P.B.; Laurie, G.W. Lacritin and other new proteins of the lacrimal functional unit. Exp. Eye Res. 2009, 88, 848–858. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wang, N.; Xie, J.; Walton, S.C.; McKown, R.L.; Raab, R.W.; Ma, P.; Beck, S.L.; Coffman, G.L.; Hussaini, I.M.; et al. Restricted epithelial proliferation by lacritin via PKCalpha-dependent NFAT and mTOR pathways. J. Cell Biol. 2006, 174, 689–700. [Google Scholar] [CrossRef]
- Wang, W.; Despanie, J.; Shi, P.; Edman-Woolcott, M.C.; Lin, Y.-A.; Cui, H.; Heur, J.M.; Fini, M.E.; Hamm-Alvarez, S.F.; MacKay, J.A. Lacritin-mediated regeneration of the corneal epithelia by protein polymer nanoparticles. J. Mater. Chem. B Mater. Biol. Med. 2014, 2, 8131–8141. [Google Scholar] [CrossRef] [Green Version]
- McKown, R.L.; Coleman Frazier, E.V.; Zadrozny, K.K.; Deleault, A.M.; Raab, R.W.; Ryan, D.S.; Sia, R.K.; Lee, J.K.; Laurie, G.W. A cleavage-potentiated fragment of tear lacritin is bactericidal. J. Biol. Chem. 2014, 289, 22172–22182. [Google Scholar] [CrossRef] [Green Version]
- Vijmasi, T.; Chen, F.Y.T.; Balasubbu, S.; Gallup, M.; McKown, R.L.; Laurie, G.W.; McNamara, N.A. Topical Administration of Lacritin Is a Novel Therapy for Aqueous-Deficient Dry Eye Disease. Investig. Ophthalmol. Vis. Sci. 2014, 55, 5401–5409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caputo, E.; Camarca, A.; Moharram, R.; Tornatore, P.; Thatcher, B.; Guardiola, J.; Martin, B.M. Structural study of GCDFP-15/gp17 in disease versus physiological conditions using a proteomic approach. Biochemistry 2003, 42, 6169–6178. [Google Scholar] [CrossRef] [PubMed]
- Chiu, W.W.-C.; Chamley, L.W. Human seminal plasma prolactin-inducible protein is an immunoglobulin G-binding protein. J. Reprod. Immunol. 2003, 60, 97–111. [Google Scholar] [CrossRef]
- Hassan, M.I.; Waheed, A.; Yadav, S.; Singh, T.P.; Ahmad, F. Prolactin inducible protein in cancer, fertility and immunoregulation: Structure, function and its clinical implications. Cell. Mol. Life Sci. 2009, 66, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Petrakis, N.L.; Lowenstein, J.M.; Wiencke, J.K.; Lee, M.M.; Wrensch, M.R.; King, E.B.; Hilton, J.F.; Miike, R. Gross cystic disease fluid protein in nipple aspirates of breast fluid of Asian and non-Asian women. Cancer Epidemiol. Biomark. Prev. 1993, 2, 573–579. [Google Scholar]
- Edechi, C.A.; Ikeogu, N.M.; Akaluka, G.N.; Terceiro, L.E.L.; Machado, M.; Salako, E.S.; Barazandeh, A.F.; Kung, S.K.P.; Uzonna, J.E.; Myal, Y. The Prolactin Inducible Protein Modulates Antitumor Immune Responses and Metastasis in a Mouse Model of Triple Negative Breast Cancer. Front. Oncol. 2021, 11, 456. [Google Scholar] [CrossRef]
- You, J.; Fitzgerald, A.; Cozzi, P.J.; Zhao, Z.; Graham, P.; Russell, P.J.; Walsh, B.J.; Willcox, M.; Zhong, L.; Wasinger, V.; et al. Post-translation modification of proteins in tears. Electrophoresis 2010, 31, 1853–1861. [Google Scholar] [CrossRef]
- Rieg, S.; Garbe, C.; Sauer, B.; Kalbacher, H.; Schittek, B. Dermcidin is constitutively produced by eccrine sweat glands and is not induced in epidermal cells under inflammatory skin conditions. Br. J. Dermatol. 2004, 151, 534–539. [Google Scholar] [CrossRef]
- Rieg, S.; Steffen, H.; Seeber, S.; Humeny, A.; Kalbacher, H.; Dietz, K.; Garbe, C.; Schittek, B. Deficiency of dermcidin-derived antimicrobial peptides in sweat of patients with atopic dermatitis correlates with an impaired innate defense of human skin in vivo. J. Immunol. 2005, 174, 8003–8010. [Google Scholar] [CrossRef] [Green Version]
- Chang, W.C.; Huang, M.S.; Yang, C.J.; Wang, W.Y.; Lai, T.C.; Hsiao, M.; Chen, C.H. Dermcidin identification from exhaled air for lung cancer diagnosis. Eur. Respir. J. 2010, 35, 1182–1185. [Google Scholar] [CrossRef]
- Stewart, G.D.; Skipworth, R.J.E.; Pennington, C.J.; Lowrie, A.G.; Deans, D.A.C.; Edwards, D.R.; Habib, F.K.; Riddick, A.C.P.; Fearon, K.C.H.; Ross, J.A. Variation in dermcidin expression in a range of primary human tumours and in hypoxic/oxidatively stressed human cell lines. Br. J. Cancer 2008, 99, 126–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zackular, J.P.; Chazin, W.J.; Skaar, E.P. Nutritional Immunity: S100 Proteins at the Host-Pathogen Interface. J. Biol. Chem. 2015, 290, 18991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donato, R.; Cannon, B.R.; Sorci, G.; Riuzzi, F.; Hsu, K.; Weber, D.J.; Geczy, C.L. Functions of S100 Proteins. Curr. Mol. Med. 2013, 13, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donato, R. Intracellular and extracellular roles of S100 proteins. Microsc. Res. Tech. 2003, 60, 540–551. [Google Scholar] [CrossRef]
- Harder, J.; Schröder, J.M. RNase 7, a novel innate immune defense antimicrobial protein of healthy human skin. J. Biol. Chem. 2002, 277, 46779–46784. [Google Scholar] [CrossRef] [Green Version]
- Rademacher, F.; Dreyer, S.; Kopfnagel, V.; Gläser, R.; Werfel, T.; Harder, J. The Antimicrobial and Immunomodulatory Function of RNase 7 in Skin. Front. Immunol. 2019, 10, 2553. [Google Scholar] [CrossRef] [Green Version]
- Spencer, J.D.; Schwaderer, A.L.; Wang, H.; Bartz, J.; Kline, J.; Eichler, T.; Desouza, K.R.; Sims-Lucas, S.; Baker, P.; Hains, D.S. Ribonuclease 7, an antimicrobial peptide upregulated during infection, contributes to microbial defense of the human urinary tract. Kidney Int. 2013, 83, 615–625. [Google Scholar] [CrossRef] [Green Version]
- Canny, G.; Levy, O. Bactericidal/permeability-increasing protein (BPI) and BPI homologs at mucosal sites. Trends Immunol. 2008, 29, 541–547. [Google Scholar] [CrossRef]
- Wang, L.; Liu, Z.; Dong, Z.; Pan, J.; Ma, X. Azurocidin-induced inhibition of oxygen metabolism in mitochondria is antagonized by heparin. Exp. Ther. Med. 2014, 8, 1473. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.; Carmona, S.; Sukhumalchandra, P.; Roszik, J.; Philips, A.; Perakis, A.A.; Kerros, C.; Zhang, M.; Qiao, N.; St. John, L.S.; et al. Cathepsin G is expressed by acute lymphoblastic leukemia and is a potential immunotherapeutic target. Front. Immunol. 2018, 8, 1975. [Google Scholar] [CrossRef]
- Melino, S.; Santone, C.; Di Nardo, P.; Sarkar, B. Histatins: Salivary peptides with copper(II)- and zinc(II)-binding motifs. FEBS J. 2014, 281, 657–672. [Google Scholar] [CrossRef] [PubMed]
- Mak, P. Hemocidins in a functional and structural context of human antimicrobial peptides. Front. Biosci. 2008, 13, 6859–6871. [Google Scholar] [CrossRef] [PubMed]
- Posta, N.; Csősz, É.; Oros, M.; Pethő, D.; Potor, L.; Kalló, G.; Hendrik, Z.; Sikura, K.É.; Méhes, G.; Tóth, C.; et al. Hemoglobin oxidation generates globin-derived peptides in atherosclerotic lesions and intraventricular hemorrhage of the brain, provoking endothelial dysfunction. Lab. Investig. 2020, 100, 986–1002. [Google Scholar] [CrossRef] [PubMed]
- Mak, P.; Wójcik, K.; Silberring, J.; Dubin, A. Antimicrobial peptides derived from heme-containing proteins: Hemocidins. Antonie Van Leeuwenhoek 2000, 77, 197–207. [Google Scholar] [CrossRef]
- Naveed, M.; Nadeem, F.; Mehmood, T.; Bilal, M.; Anwar, Z.; Amjad, F. Protease—A Versatile and Ecofriendly Biocatalyst with Multi-Industrial Applications: An Updated Review. Catal. Lett. 2021, 151, 307–323. [Google Scholar] [CrossRef]
- Feng, Y.; Li, Q.; Chen, J.; Yi, P.; Xu, X.; Fan, Y.; Cui, B.; Yu, Y.; Li, X.; Du, Y.; et al. Salivary protease spectrum biomarkers of oral cancer. Int. J. Oral Sci. 2019, 11, 7. [Google Scholar] [CrossRef]
- Fu, R.; Klinngam, W.; Heur, M.; Edman, M.C.; Hamm-Alvarez, S.F. Tear proteases and protease inhibitors: Potential biomarkers and disease drivers in ocular surface disease. Eye Contact Lens 2020, 46, S70. [Google Scholar] [CrossRef]
- Magalhães, B.; Trindade, F.; Barros, A.S.; Klein, J.; Amado, F.; Ferreira, R.; Vitorino, R. Reviewing Mechanistic Peptidomics in Body Fluids Focusing on Proteases. Proteomics 2018, 18, 1800187. [Google Scholar] [CrossRef]
- Vandooren, J.; Itoh, Y. Alpha-2-Macroglobulin in Inflammation, Immunity and Infections. Front. Immunol. 2021, 12, 5411. [Google Scholar] [CrossRef]
- Ranasinghe, S.L.; McManus, D.P. Protease Inhibitors of Parasitic Flukes: Emerging Roles in Parasite Survival and Immune Defence. Trends Parasitol. 2017, 33, 400–413. [Google Scholar] [CrossRef]
- Janciauskiene, S.; Wrenger, S.; Immenschuh, S.; Olejnicka, B.; Greulich, T.; Welte, T.; Chorostowska-Wynimko, J. The multifaceted effects of Alpha1-Antitrypsin on neutrophil functions. Front. Pharmacol. 2018, 9, 341. [Google Scholar] [CrossRef] [PubMed]
- Roemisch, J.; Gray, E.; Hoffmann, J.N.; Wiedermann, C.J.; Kalina, U. Antithrombin: A new look at the actions of a serine protease inhibitor. Blood Coagul. Fibrinolysis 2002, 13, 657–670. [Google Scholar] [CrossRef] [PubMed]
- Agbowuro, A.A.; Huston, W.M.; Gamble, A.B.; Tyndall, J.D.A. Proteases and protease inhibitors in infectious diseases. Med. Res. Rev. 2018, 38, 1295–1331. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Gao, J.; Tang, W. Nosocomial infection and its molecular mechanisms of antibiotic resistance. Biosci. Trends 2016, 10, 14–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sousa, S.A.; Feliciano, J.R.; Pita, T.; Soeiro, C.F.; Mendes, B.L.; Alves, L.G.; Leitão, J.H. Bacterial Nosocomial Infections: Multidrug Resistance as a Trigger for the Development of Novel Antimicrobials. Antibiotics 2021, 10, 942. [Google Scholar] [CrossRef]
- Wieler, L.H.; Ewers, C.; Guenther, S.; Walther, B.; Lübke-Becker, A. Methicillin-resistant staphylococci (MRS) and extended-spectrum beta-lactamases (ESBL)-producing Enterobacteriaceae in companion animals: Nosocomial infections as one reason for the rising prevalence of these potential zoonotic pathogens in clinical samples. Int. J. Med. Microbiol. 2011, 301, 635–641. [Google Scholar] [CrossRef]
- Nguyen, M.; Joshi, S.G. Carbapenem resistance in Acinetobacter baumannii, and their importance in hospital-acquired infections: A scientific review. J. Appl. Microbiol. 2021, 131, 2715–2738. [Google Scholar] [CrossRef]
- Mak, S.; Xu, Y.; Nodwell, J.R. The expression of antibiotic resistance genes in antibiotic-producing bacteria. Mol. Microbiol. 2014, 93, 391–402. [Google Scholar] [CrossRef]
- Avershina, E.; Shapovalova, V.; Shipulin, G. Fighting Antibiotic Resistance in Hospital-Acquired Infections: Current State and Emerging Technologies in Disease Prevention, Diagnostics and Therapy. Front. Microbiol. 2021, 12, 2044. [Google Scholar] [CrossRef]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The value of antimicrobial peptides in the age of resistance. Lancet. Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef]
- Kosikowska, P.; Lesner, A. Antimicrobial peptides (AMPs) as drug candidates: A patent review (2003–2015). Expert Opin. Ther. Pat. 2016, 26, 689–702. [Google Scholar] [CrossRef] [PubMed]
- Wach, A.; Dembowsky, K.; Dale, G.E. Pharmacokinetics and Safety of Intravenous Murepavadin Infusion in Healthy Adult Subjects Administered Single and Multiple Ascending Doses. Antimicrob. Agents Chemother. 2018, 62, e02355-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otvos, L.; Ostorhazi, E.; Szabo, D.; Zumbrun, S.D.; Miller, L.L.; Halasohoris, S.A.; Desai, P.D.; Veldt, S.M.I.; Kraus, C.N. Synergy Between Proline-Rich Antimicrobial Peptides and Small Molecule Antibiotics Against Selected Gram-Negative Pathogens in vitro and in vivo. Front. Chem. 2018, 6, 309. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Bharath Prasad, A.S.; Mehta, C.H.; Nayak, U.Y. Antimicrobial peptide polymers: No escape to ESKAPE pathogens—A review. World J. Microbiol. Biotechnol. 2020, 36, 131. [Google Scholar] [CrossRef] [PubMed]
- Van der Plas, M.J.A.; Cai, J.; Petrlova, J.; Saleh, K.; Kjellström, S.; Schmidtchen, A. Method development and characterisation of the low-molecular-weight peptidome of human wound fluids. Elife 2021, 10, e66876. [Google Scholar] [CrossRef]
- Hartman, E.; Wallblom, K.; van der Plas, M.J.A.; Petrlova, J.; Cai, J.; Saleh, K.; Kjellström, S.; Schmidtchen, A. Bioinformatic Analysis of the Wound Peptidome Reveals Potential Biomarkers and Antimicrobial Peptides. Front. Immunol. 2021, 11, 620707. [Google Scholar] [CrossRef]
- Dahlman, A.; Puthia, M.; Petrlova, J.; Schmidtchen, A.; Petruk, G. Thrombin-Derived C-Terminal Peptide Reduces Candida-Induced Inflammation and Infection In Vitro and In Vivo. Antimicrob. Agents Chemother. 2021, 65, e01032-21. [Google Scholar] [CrossRef]
- Hu, S.; Loo, J.A.; Wong, D.T. Human body fluid proteome analysis. Proteomics 2006, 6, 6326. [Google Scholar] [CrossRef]
- Leeman, M.; Choi, J.; Hansson, S.; Storm, M.U.; Nilsson, L. Proteins and antibodies in serum, plasma, and whole blood—size characterization using asymmetrical flow field-flow fractionation (AF4). Anal. Bioanal. Chem. 2018, 410, 4867. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Shao, D.; Wang, Y.; Cui, X.; Li, Y.; Chen, Q.; Cui, J. Human body-fluid proteome: Quantitative profiling and computational prediction. Brief. Bioinform. 2021, 22, 315–333. [Google Scholar] [CrossRef] [Green Version]
- Bellei, E.; Bergamini, S.; Monari, E.; Fantoni, L.I.; Cuoghi, A.; Ozben, T.; Tomasi, A. High-abundance proteins depletion for serum proteomic analysis: Concomitant removal of non-targeted proteins. Amino Acids 2011, 40, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Chien, S.C.; Chen, C.Y.; Lin, C.F.; Yeh, H.I. Critical appraisal of the role of serum albumin in cardiovascular disease. Biomark. Res. 2017, 5, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, S.; Gautam, V.; Naseem, S. Acute-phase proteins: As diagnostic tool. J. Pharm. Bioallied Sci. 2011, 3, 118. [Google Scholar] [CrossRef] [PubMed]
- Gum, E.T.; Swanson, R.A.; Alano, C.; Liu, J.; Hong, S.; Weinstein, P.R.; Panter, S.S. Human serum albumin and its N-terminal tetrapeptide (DAHK) block oxidant-induced neuronal death. Stroke 2004, 35, 590–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomme, P.T.; McCann, K.B. Transferrin: Structure, function and potential therapeutic actions. Drug Discov. Today 2005, 10, 267–273. [Google Scholar] [CrossRef]
- Von Bonsdorff, L.; Sahlstedt, L.; Ebeling, F.; Ruutu, T.; Parkkinen, J. Apotransferrin administration prevents growth of Staphylococcus epidermidis in serum of stem cell transplant patients by binding of free iron. FEMS Immunol. Med. Microbiol. 2003, 37, 45–51. [Google Scholar] [CrossRef] [Green Version]
- Ardehali, R.; Shi, L.; Janatova, J.; Mohammad, S.F.; Burns, G.L. The inhibitory activity of serum to prevent bacterial adhesion is mainly due to apo-transferrin. J. Biomed. Mater. Res. A 2003, 66, 21–28. [Google Scholar] [CrossRef]
- Alayash, A.I.; Andersen, C.B.F.; Moestrup, S.K.; Bülow, L. Haptoglobin: The hemoglobin detoxifier in plasma. Trends Biotechnol. 2013, 31, 2–3. [Google Scholar] [CrossRef]
- Kristiansen, M.; Graversen, J.H.; Jacobsen, C.; Sonne, O.; Hoffman, H.J.; Law, S.K.A.; Moestrup, S.K. Identification of the haemoglobin scavenger receptor. Nature 2001, 409, 198–201. [Google Scholar] [CrossRef]
- MacKellar, M.; Vigerust, D.J. Role of Haptoglobin in Health and Disease: A Focus on Diabetes. Clin. Diabetes 2016, 34, 148. [Google Scholar] [CrossRef] [Green Version]
- Dominiczak, M.H.; Caslake, M.J. Apolipoproteins: Metabolic role and clinical biochemistry applications. Ann. Clin. Biochem. 2011, 48, 498–515. [Google Scholar] [CrossRef] [PubMed]
- Tada, N.; Sakamoto, T.; Kagami, A.; Mochizuki, K.; Kurosaka, K. Antimicrobial activity of lipoprotein particles containing apolipoprotein Al. Mol. Cell. Biochem. 1993, 119, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Dell’Olmo, E.; Gaglione, R.; Sabbah, M.; Schibeci, M.; Cesaro, A.; Di Girolamo, R.; Porta, R.; Arciello, A. Host defense peptides identified in human apolipoprotein B as novel food biopreservatives and active coating components. Food Microbiol. 2021, 99, 103804. [Google Scholar] [CrossRef] [PubMed]
- Nokhoijav, E.; Guba, A.; Kumar, A.; Kunkli, B.; Kalló, G.; Káplár, M.; Somodi, S.; Garai, I.; Csutak, A.; Tóth, N.; et al. Metabolomic Analysis of Serum and Tear Samples from Patients with Obesity and Type 2 Diabetes Mellitus. Int. J. Mol. Sci. 2022, 23, 4534. [Google Scholar] [CrossRef]
- Prabha, J.L. Tear Secretion-A Short Review. J. Pharm. Sci. Res. 2014, 6, 155–157. [Google Scholar]
- Fullard, R.J.; Snyder, C. Protein levels in nonstimulated and stimulated tears of normal human subjects. Investig. Ophthalmol. Vis. Sci. 1990, 31, 1119–1126. [Google Scholar]
- Tiffany, J.M. Tears in health and disease. Eye 2003, 17, 923–926. [Google Scholar] [CrossRef]
- Janeway, J.C.A.; Travers, P.; Walport, M.; Shlomchik, M.J. The Front Line of Host Defense, 5th ed.; Garland Science: New York, NY, USA, 2001. [Google Scholar]
- Shao, D.; Huang, L.; Wang, Y.; Cui, X.; Li, Y.; Wang, Y.; Ma, Q.; Du, W.; Cui, J. HBFP: A new repository for human body fluid proteome. Database 2021, 2021, baab065. [Google Scholar] [CrossRef]
- Janssen, P.T.; van Bijsterveld, O.P. Origin and biosynthesis of human tear fluid proteins. Investig. Ophthalmol. Vis. Sci. 1983, 24, 623–630. [Google Scholar]
- Humphrey, S.P.; Williamson, R.T. A review of saliva: Normal composition, flow, and function. J. Prosthet. Dent. 2001, 85, 162–169. [Google Scholar] [CrossRef]
- Shaila, M.; Pai, G.P.; Shetty, P. Salivary protein concentration, flow rate, buffer capacity and pH estimation: A comparative study among young and elderly subjects, both normal and with gingivitis and periodontitis. J. Indian Soc. Periodontol. 2013, 17, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Henskens, Y.M.; van der Velden, U.; Veerman, E.C.; Nieuw Amerongen, A.V. Protein, albumin and cystatin concentrations in saliva of healthy subjects and of patients with gingivitis or periodontitis. J. Periodontal Res. 1993, 28, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Schulz, B.L.; Cooper-White, J.; Punyadeera, C.K. Saliva proteome research: Current status and future outlook. Crit. Rev. Biotechnol. 2013, 33, 246–259. [Google Scholar] [CrossRef] [PubMed]
- Csősz, É.; Kalló, G.; Márkus, B.; Deák, E.; Csutak, A.; Tőzsér, J. Quantitative body fluid proteomics in medicine—A focus on minimal invasiveness. J. Proteom. 2016, 153, 30364–30365. [Google Scholar] [CrossRef] [Green Version]
- Lahiri, D.; Nag, M.; Banerjee, R.; Mukherjee, D.; Garai, S.; Sarkar, T.; Dey, A.; Sheikh, H.I.; Pathak, S.K.; Edinur, H.A.; et al. Amylases: Biofilm Inducer or Biofilm Inhibitor? Front. Cell. Infect. Microbiol. 2021, 11, 355. [Google Scholar] [CrossRef]
- Bechler, J.; Bermudez, L.E. Investigating the Role of Mucin as Frontline Defense of Mucosal Surfaces against Mycobacterium avium Subsp. hominissuis. J. Pathog. 2020, 2020, 9451591. [Google Scholar] [CrossRef]
- Linden, S.K.; Sutton, P.; Karlsson, N.G.; Korolik, V.; McGuckin, M.A. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008, 1, 183–197. [Google Scholar] [CrossRef] [Green Version]
- Eaves-Pyles, T.; Patel, J.; Arigi, E.; Cong, Y.; Cao, A.; Garg, N.; Dhiman, M.; Pyles, R.B.; Arulanandam, B.; Miller, A.L.; et al. Immunomodulatory and antibacterial effects of cystatin 9 against Francisella tularensis. Mol. Med. 2013, 19, 263–275. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Russell, M.W. Innate Humoral Defense Factors. Mucosal Immunol. Fourth Ed. 2015, 1–2, 251–270. [Google Scholar] [CrossRef]
- Levine, M. Susceptibility to Dental Caries and the Salivary Proline-Rich Proteins. Int. J. Dent. 2011, 2011, 953412. [Google Scholar] [CrossRef] [Green Version]
- Metz-Boutigue, M.-H.; Shooshtarizadeh, P.; Prevost, G.; Haikel, Y.; Chich, J.-F. Antimicrobial peptides present in mammalian skin and gut are multifunctional defence molecules. Curr. Pharm. Des. 2010, 16, 1024–1039. [Google Scholar] [CrossRef] [PubMed]
- ROBINSON, S.; ROBINSON, A.H. Chemical composition of sweat. Physiol. Rev. 1954, 34, 202–220. [Google Scholar] [CrossRef] [PubMed]
- Harder, J.; Schröder, J.-M.; Gläser, R. The skin surface as antimicrobial barrier: Present concepts and future outlooks. Exp. Dermatol. 2013, 22, 1–5. [Google Scholar] [CrossRef]
- Park, J.H.; Park, G.T.; Cho, I.H.; Sim, S.M.; Yang, J.M.; Lee, D.Y. An antimicrobial protein, lactoferrin exists in the sweat: Proteomic analysis of sweat. Exp. Dermatol. 2011, 20, 369–371. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.; Ledee, D.R.; Gordon, G.M.; Itakura, T.; Patel, N.; Martin, A.; Fini, M.E. Interaction of clusterin and matrix metalloproteinase-9 and its implication for epithelial homeostasis and inflammation. Am. J. Pathol. 2012, 180, 2028–2039. [Google Scholar] [CrossRef] [Green Version]
- Rizzi, F.; Bettuzzi, S. The clusterin paradigm in prostate and breast carcinogenesis. Endocr. Relat. Cancer 2010, 17, R1–R17. [Google Scholar] [CrossRef] [Green Version]
- Sueno, K.; Nakaima, N.; Shingaki, K.; Ura, M.; Noda, Y.; Kosugi, T.; Eichner, H. Total protein concentration in selectively collected secretions from the middle and inferior meatus of the nose. Auris. Nasus. Larynx 1986, 13 (Suppl. 1), S85–S88. [Google Scholar] [CrossRef]
- Ruocco, L.; Fattori, B.; Romanelli, A.; Martelloni, M.; Casani, A.; Samolewska, M.; Rezzonico, R. A new collection method for the evaluation of nasal mucus proteins. Clin. Exp. Allergy 1998, 28, 881–888. [Google Scholar] [CrossRef]
- Cole, A.M.; Dewan, P.; Ganz, T. Innate antimicrobial activity of nasal secretions. Infect. Immun. 1999, 67, 3267–3275. [Google Scholar] [CrossRef] [Green Version]
- Meredith, S.D.; Raphael, G.D.; Baraniuk, J.N.; Banks, S.M.; Kaliner, M.A. The pathophysiology of rhinitis. III. The control of IgG secretion. J. Allergy Clin. Immunol. 1989, 84, 920–930. [Google Scholar] [CrossRef]
- Saieg, A.; Brown, K.J.; Pena, M.T.; Rose, M.C.; Preciado, D. Proteomic analysis of pediatric sinonasal secretions shows increased MUC5B mucin in CRS. Pediatr. Res. 2015, 77, 356–362. [Google Scholar] [CrossRef]
- Casado, B.; Pannell, L.K.; Viglio, S.; Iadarola, P.; Baraniuk, J.N. Analysis of the sinusitis nasal lavage fluid proteome using capillary liquid chromatography interfaced to electrospray ionization-quadrupole time of flight- tandem mass spectrometry. Electrophoresis 2004, 25, 1386–1393. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chavali, S.; Mobini, R.; Muraro, A.; Barbon, F.; Boldrin, D.; Åberg, N.; Benson, M. A pathway-based approach to find novel markers of local glucocorticoid treatment in intermittent allergic rhinitis. Allergy Eur. J. Allergy Clin. Immunol. 2011, 66, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Tomazic, P.V.; Darnhofer, B.; Birner-Gruenberger, R. Nasal mucus proteome and its involvement in allergic rhinitis. Expert Rev. Proteom. 2020, 17, 191–199. [Google Scholar] [CrossRef] [Green Version]
- Brunzel, N.A. Fundamentals of Urine and Body Fluid Analysis, 3rd ed.; Elsevier/Saunders: St. Louis, MO, USA, 2013. [Google Scholar]
- Zhao, M.; Li, M.; Yang, Y.; Guo, Z.; Sun, Y.; Shao, C.; Li, M.; Sun, W.; Gao, Y. A comprehensive analysis and annotation of human normal urinary proteome. Sci. Rep. 2017, 7, 3024. [Google Scholar] [CrossRef]
- Devuyst, O.; Olinger, E.; Rampoldi, L. Uromodulin: From physiology to rare and complex kidney disorders. Nat. Rev. Nephrol. 2017, 13, 525–544. [Google Scholar] [CrossRef]
- Weiss, G.L.; Stanisich, J.J.; Sauer, M.M.; Lin, C.W.; Eras, J.; Zyla, D.S.; Trück, J.; Devuyst, O.; Aebi, M.; Pilhofer, M.; et al. Architecture and function of human uromodulin filaments in urinary tract infections. Science 2020, 369, 1005–1010. [Google Scholar] [CrossRef]
- Kukulski, W. A glycoprotein in urine binds bacteria and blocks infections. Science 2020, 369, 917–918. [Google Scholar] [CrossRef]
- Wu, T.H.; Li, K.J.; Yu, C.L.; Tsai, C.Y. Tamm–Horsfall Protein is a Potent Immunomodulatory Molecule and a Disease Biomarker in the Urinary System. Molecules 2018, 23, 200. [Google Scholar] [CrossRef] [Green Version]
- Bergwik, J.; Kristiansson, A.; Allhorn, M.; Gram, M.; Åkerström, B. Structure, Functions, and Physiological Roles of the Lipocalin α1-Microglobulin (A1M). Front. Physiol. 2021, 12, 251. [Google Scholar] [CrossRef]
- Åkerström, B.; Gram, M. A1M, an extravascular tissue cleaning and housekeeping protein. Free Radic. Biol. Med. 2014, 74, 274–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gunnarsson, R.; Åkerström, B.; Hansson, S.R.; Gram, M. Recombinant alpha-1-microglobulin: A potential treatment for preeclampsia. Drug Discov. Today 2017, 22, 736–743. [Google Scholar] [CrossRef] [PubMed]
- Kristiansson, A.; Gram, M.; Flygare, J.; Hansson, S.R.; Åkerström, B.; Storry, J.R. The Role of α 1-Microglobulin (A1M) in Erythropoiesis and Erythrocyte Homeostasis-Therapeutic Opportunities in Hemolytic Conditions. Int. J. Mol. Sci. 2020, 21, 7234. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Calero, L.; Martin-Lorenzo, M.; Ramos-Barron, A.; Ruiz-Criado, J.; Maroto, A.S.; Ortiz, A.; Gomez-Alamillo, C.; Arias, M.; Vivanco, F.; Alvarez-Llamas, G. Urinary Kininogen-1 and Retinol binding protein-4 respond to Acute Kidney Injury: Predictors of patient prognosis? Sci. Rep. 2016, 6, 19667. [Google Scholar] [CrossRef] [Green Version]
- Wong, M.K.-S. Kininogen; Academic Press: Cambridge, MA, USA, 2021. [Google Scholar]
- Weinberg, M.S.; Azar, P.; Trebbin, W.M.; Solomon, R.J. The role of urinary kininogen in the regulation of kinin generation. Kidney Int. 1985, 28, 975–981. [Google Scholar] [CrossRef] [Green Version]
- Ben Nasr, A.; Herwald, H.; Muller-Esterl, W.; Bjorck, L. Human kininogens interact with M protein, a bacterial surface protein and virulence determinant. Biochem. J. 1995, 305, 173–180. [Google Scholar] [CrossRef] [Green Version]
- Rapala-Kozik, M.; Karkowska, J.; Jacher, A.; Golda, A.; Barbasz, A.; Guevara-Lora, I.; Kozik, A. Kininogen adsorption to the cell surface of Candida spp. Int. Immunopharmacol. 2008, 8, 237–241. [Google Scholar] [CrossRef]
- Sonesson, A.; Nordahl, E.A.; Malmsten, M.; Schmidtchen, A. Antifungal activities of peptides derived from domain 5 of high-molecular-weight kininogen. Int. J. Pept. 2011, 2011, 1–12. [Google Scholar] [CrossRef]
- Hitti, J.; Lapidus, J.A.; Lu, X.; Reddy, A.P.; Jacob, T.; Dasari, S.; Eschenbach, D.A.; Gravett, M.G.; Nagalla, S.R. Noninvasive diagnosis of intraamniotic infection: Proteomic biomarkers in vaginal fluid. Am. J. Obstet. Gynecol. 2010, 203, 32.e1–32.e8. [Google Scholar] [CrossRef] [Green Version]
- Pizzorno, J.E.; Murray, M.T.; Joiner-Bey, H. Vaginitis, 3rd ed.; Churchill Livingstone: London, UK, 2016; ISBN 978-0-7020-5514-0. [Google Scholar]
- Zegels, G.; Van Raemdonck, G.A.A.; Tjalma, W.A.A.; Van Ostade, X.W.M. Use of cervicovaginal fluid for the identification of biomarkers for pathologies of the female genital tract. Proteome Sci. 2010, 8, 63. [Google Scholar] [CrossRef] [Green Version]
- Starodubtseva, N.L.; Brzhozovskiy, A.G.; Bugrova, A.E.; Kononikhin, A.S.; Indeykina, M.I.; Gusakov, K.I.; Chagovets, V.V.; Nazarova, N.M.; Frankevich, V.E.; Sukhikh, G.T.; et al. Label-free cervicovaginal fluid proteome profiling reflects the cervix neoplastic transformation. J. Mass Spectrom. 2019, 54, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Muytjens, C.M.J.; Yu, Y.; Diamandis, E.P. Discovery of Antimicrobial Peptides in Cervical-Vaginal Fluid from Healthy Nonpregnant Women via an Integrated Proteome and Peptidome Analysis. Proteomics 2017, 17, 1600461. [Google Scholar] [CrossRef] [PubMed]
- Schjenken, J.E.; Robertson, S.A. The Female Response to Seminal Fluid. Physiol. Rev. 2020, 100, 1077–1117. [Google Scholar] [CrossRef] [PubMed]
- Schjenken, J.E.; Robertson, S.A. Seminal fluid and immune adaptation for pregnancy--comparative biology in mammalian species. Reprod. Domest. Anim. 2014, 49 (Suppl. 3), 27–36. [Google Scholar] [CrossRef]
- Pilch, B.; Mann, M. Large-scale and high-confidence proteomic analysis of human seminal plasma. Genome Biol. 2006, 7, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Klavert, J.; van der Eerden, B.C.J. Fibronectin in Fracture Healing: Biological Mechanisms and Regenerative Avenues. Front. Bioeng. Biotechnol. 2021, 9, 274. [Google Scholar] [CrossRef]
- Speziale, P.; Arciola, C.R.; Pietrocola, G. Fibronectin and Its Role in Human Infective Diseases. Cells 2019, 8, 1516. [Google Scholar] [CrossRef] [Green Version]
- Hohenester, E. Structural biology of laminins. Essays Biochem. 2019, 63, 285–295. [Google Scholar] [CrossRef] [Green Version]
- Bosler, J.S.; Davies, K.P.; Neal-Perry, G.S. Peptides in seminal fluid and their role in infertility: A potential role for opiorphin inhibition of neutral endopeptidase activity as a clinically relevant modulator of sperm motility: A review. Reprod. Sci. 2014, 21, 1334–1340. [Google Scholar] [CrossRef] [Green Version]
- Lundwall, Å.; Bjartell, A.; Olsson, A.Y.; Malm, J. Semenogelin I and II, the predominant human seminal plasma proteins, are also expressed in non-genital tissues. Mol. Hum. Reprod. 2002, 8, 805–810. [Google Scholar] [CrossRef] [Green Version]
- De Lamirande, E. Semenogelin, the main protein of the human semen coagulum, regulates sperm function. Semin. Thromb. Hemost. 2007, 33, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Lee, W.H.; Shen, J.H.; Li, H.; Zhang, Y. Identification of novel semenogelin I-derived antimicrobial peptide from liquefied human seminal plasma. Peptides 2008, 29, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Bourgeon, F.; Evrard, B.; Brillard-Bourdet, M.; Colleu, D.; Jégou, B.; Pineau, C. Involvement of semenogelin-derived peptides in the antibacterial activity of human seminal plasma. Biol. Reprod. 2004, 70, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Sakka, L.; Coll, G.; Chazal, J. Anatomy and physiology of cerebrospinal fluid. Eur. Ann. Otorhinolaryngol. Head Neck Dis. 2011, 128, 309–316. [Google Scholar] [CrossRef] [Green Version]
- McPherson RA, P.M. Henry’s Clinical Diagnosis and Management by Laboratory Methods, 23rd ed.; W.B. Saunders: Philadelphia, PA, USA, 2006. [Google Scholar]
- McComb, J.G. Recent research into the nature of cerebrospinal fluid formation and absorption. J. Neurosurg. 1983, 59, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Begcevic, I.; Brinc, D.; Drabovich, A.P.; Batruch, I.; Diamandis, E.P. Identification of brain-enriched proteins in the cerebrospinal fluid proteome by LC-MS/MS profiling and mining of the Human Protein Atlas. Clin. Proteom. 2016, 13, 11. [Google Scholar] [CrossRef] [Green Version]
- Jankovska, E.; Svitek, M.; Holada, K.; Petrak, J. Affinity depletion versus relative protein enrichment: A side-by-side comparison of two major strategies for increasing human cerebrospinal fluid proteome coverage. Clin. Proteom. 2019, 16, 9. [Google Scholar] [CrossRef] [Green Version]
- Redzic, Z.B.; Preston, J.E.; Duncan, J.A.; Chodobski, A.; Szmydynger-Chodobska, J. The choroid plexus-cerebrospinal fluid system: From development to aging. Curr. Top. Dev. Biol. 2005, 71, 1–52. [Google Scholar] [CrossRef]
- Reiber, H. Dynamics of brain-derived proteins in cerebrospinal fluid. Clin. Chim. Acta. 2001, 310, 173–186. [Google Scholar] [CrossRef] [Green Version]
- Liz, M.A.; Coelho, T.; Bellotti, V.; Fernandez-Arias, M.I.; Mallaina, P.; Obici, L. A Narrative Review of the Role of Transthyretin in Health and Disease. Neurol. Ther. 2020, 9, 395–402. [Google Scholar] [CrossRef]
- Sharma, M.; Khan, S.; Rahman, S.; Singh, L.R. The extracellular protein, transthyretin is an oxidative stress biomarker. Front. Physiol. 2019, 10, 5. [Google Scholar] [CrossRef] [PubMed]
- Jain, N.; Ådén, J.; Nagamatsu, K.; Evans, M.L.; Li, X.; McMichael, B.; Ivanova, M.I.; Almqvist, F.; Buxbaum, J.N.; Chapman, M.R. Inhibition of curli assembly and Escherichia coli biofilm formation by the human systemic amyloid precursor transthyretin. Proc. Natl. Acad. Sci. USA 2017, 114, 12184–12189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunt, J.M.; Tuder, R. Alpha 1 anti-trypsin: One protein, many functions. Curr. Mol. Med. 2012, 12, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Janciauskiene, S.M.; Bals, R.; Koczulla, R.; Vogelmeier, C.; Köhnlein, T.; Welte, T. The discovery of α1-antitrypsin and its role in health and disease. Respir. Med. 2011, 105, 1129–1139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urade, Y. Biochemical and Structural Characteristics, Gene Regulation, Physiological, Pathological and Clinical Features of Lipocalin-Type Prostaglandin D2 Synthase as a Multifunctional Lipocalin. Front. Physiol. 2021, 12, 1627. [Google Scholar] [CrossRef]
- Ricciotti, E.; Fitzgerald, G.A. Prostaglandins and inflammation. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 986–1000. [Google Scholar] [CrossRef]
- Kanda, N.; Ishikawa, T.; Watanabe, S. Prostaglandin D2 induces the production of human beta-defensin-3 in human keratinocytes. Biochem. Pharmacol. 2010, 79, 982–989. [Google Scholar] [CrossRef]
- Guo, T.; Noble, W.; Hanger, D.P. Roles of tau protein in health and disease. Acta Neuropathol. 2017, 133, 665. [Google Scholar] [CrossRef] [Green Version]
- Michalicova, A.; Majerova, P.; Kovac, A. Tau Protein and Its Role in Blood–Brain Barrier Dysfunction. Front. Mol. Neurosci. 2020, 13, 178. [Google Scholar] [CrossRef]
- Kobayashi, N.; Masuda, J.; Kudoh, J.; Shimizu, N.; Yoshida, T. Binding sites on tau proteins as components for antimicrobial peptides. Biocontrol Sci. 2008, 13, 49–56. [Google Scholar] [CrossRef] [Green Version]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalló, G.; Kumar, A.; Tőzsér, J.; Csősz, É. Chemical Barrier Proteins in Human Body Fluids. Biomedicines 2022, 10, 1472. https://doi.org/10.3390/biomedicines10071472
Kalló G, Kumar A, Tőzsér J, Csősz É. Chemical Barrier Proteins in Human Body Fluids. Biomedicines. 2022; 10(7):1472. https://doi.org/10.3390/biomedicines10071472
Chicago/Turabian StyleKalló, Gergő, Ajneesh Kumar, József Tőzsér, and Éva Csősz. 2022. "Chemical Barrier Proteins in Human Body Fluids" Biomedicines 10, no. 7: 1472. https://doi.org/10.3390/biomedicines10071472
APA StyleKalló, G., Kumar, A., Tőzsér, J., & Csősz, É. (2022). Chemical Barrier Proteins in Human Body Fluids. Biomedicines, 10(7), 1472. https://doi.org/10.3390/biomedicines10071472