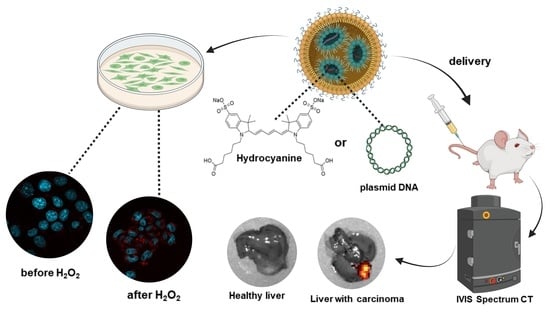
Delivery of Lipid Nanoparticles with ROS Probes for Improved Visualization of Hepatocellular Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Synthesis of Hydro-Cy5-siRNA-LNPs
2.3. Synthesis of LNPs with Plasmid Encoding HyPer7
2.4. Characterization of Obtained LNP
2.5. Cytotoxicity of Obtained Lipid Nanoparticles (MTT Assay)
2.6. Analysis of ROS Activity In Vitro
2.7. Delivery of LNPs to the Liver and Analysis of ROS Activity in Vivo in HCC Model
2.8. Statistical Analysis
3. Results
3.1. Synthesis and Characterisation of Lipid Nanoparticles and ROS Analysis In Vitro
3.2. Intravital Delivery of ROS Sensor-Lipid Nanoparticles and Visualization of Hepatocellular Carcinoma
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zou, Z.; Chang, H.; Li, H.; Wang, S. Induction of Reactive Oxygen Species: An Emerging Approach for Cancer Therapy. Apoptosis 2017, 22, 1321–1335. [Google Scholar] [CrossRef]
- Trachootham, D.; Alexandre, J.; Huang, P. Targeting Cancer Cells by ROS-Mediated Mechanisms: A Radical Therapeutic Approach? Nat. Rev. Drug Discov. 2009, 8, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Boonstra, J.; Post, J.A. Molecular Events Associated with Reactive Oxygen Species and Cell Cycle Progression in Mammalian Cells. Gene 2004, 337, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Ji, Y.; Li, X.; Ding, J.; Chen, L.; Huang, Y.; Wei, W. Uri1 Suppresses Irradiation-Induced Reactive Oxygen Species (Ros) by Activating Autophagy in Hepatocellular Carcinoma Cells. Int. J. Biol. Sci. 2021, 17, 3091–3103. [Google Scholar] [CrossRef] [PubMed]
- Cichoz-Lach, H.; Michalak, A. Oxidative Stress as a Crucial Factor in Liver Diseases. World J. Gastroenterol. 2014, 20, 8082–8091. [Google Scholar] [CrossRef]
- Rezende, F.; Brandes, R.P.; Schröder, K. Detection of Hydrogen Peroxide with Fluorescent Dyes. Antioxid. Redox. Signal. 2018, 29, 585–602. [Google Scholar] [CrossRef] [PubMed]
- Abakumova, T.; Prikazchikova, T.; Aparin, I.; Vaneev, A.; Gorelkin, P.; Erofeev, A.; Zatsepin, T. ROS-Sensitive Dyes in Lipid Nanoparticles for in Vivo Imaging. In Proceedings of the 2020 IEEE 10th International Conference on “Nanomaterials: Applications and Properties”, NAP 2020, Sumy, Ukraine, 9–13 November 2020; Institute of Electrical and Electronics Engineers Inc.: Piscataway, NJ, USA, 2020. [Google Scholar]
- Wang, H.; Wang, X.; Li, P.; Dong, M.; Yao, S.Q.; Tang, B. Fluorescent Probes for Visualizing ROS-Associated Proteins in Disease. Chem. Sci. 2021, 12, 11620–11646. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Dai, M.; Yuan, Z. Methods for the Detection of Reactive Oxygen Species. Anal. Methods 2018, 10, 4625–4638. [Google Scholar] [CrossRef]
- Bai, X.; King-Hei Ng, K.; Jacob Hu, J.; Ye, S.; Yang, D. Small-Molecule-Based Fluorescent Sensors for Selective Detection of Reactive Oxygen Species in Biological Systems. Annu. Rev. Biochem. 2019, 88, 605–633. [Google Scholar] [CrossRef] [PubMed]
- Oushiki, D.; Kojima, H.; Terai, T.; Arita, M.; Hanaoka, K.; Urano, Y.; Nagano, T. Development and Application of a Near-Infrared Fluorescence Probe for Oxidative Stress Based on Differential Reactivity of Linked Cyanine Dyes. J. Am. Chem. Soc. 2010, 132, 2795–2801. [Google Scholar] [CrossRef]
- Wardman, P. Fluorescent and Luminescent Probes for Measurement of Oxidative and Nitrosative Species in Cells and Tissues: Progress, Pitfalls, and Prospects. Free Radic. Biol. Med. 2007, 43, 995–1022. [Google Scholar] [CrossRef]
- Winterbourn, C.C. The Challenges of Using Fluorescent Probes to Detect and Quantify Specific Reactive Oxygen Species in Living Cells. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 730–738. [Google Scholar] [CrossRef]
- Kim, J.Y.; Choi, W.I.; Kim, Y.H.; Tae, G. Highly Selective In-Vivo Imaging of Tumor as an Inflammation Site by ROS Detection Using Hydrocyanine-Conjugated, Functional Nano-Carriers. J. Control. Release 2011, 156, 398–405. [Google Scholar] [CrossRef] [PubMed]
- Erard, M.; Dupré-Crochet, S.; Nüße, X.O. Biosensors for Spatiotemporal Detection of Reactive Oxygen Species in Cells and Tissues. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2018, 314, 667–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, H.; Aleyasin, H.; Dickinson, B.C.; Haskew-Layton, R.E.; Ratan, R.R. Recent Advances in Hydrogen Peroxide Imaging for Biological Applications. Cell Biosci. 2014, 4, 64. [Google Scholar] [CrossRef] [Green Version]
- Kim, G.; Koo, Y.E.L.; Xu, H.; Philbert, M.A.; Kopelman, R. Nanoencapsulation Method for High Selectivity Sensing of Hydrogen Peroxide inside Live Cells. Anal. Chem. 2010, 82, 2165–2169. [Google Scholar] [CrossRef] [Green Version]
- Hammond, V.J.; Aylott, J.W.; Greenway, G.M.; Watts, P.; Webster, A.; Wiles, C. An Optical Sensor for Reactive Oxygen Species: Encapsulation of Functionalised Silica Nanoparticles into Silicate Nanoprobes to Reduce Fluorophore Leaching. Analyst 2008, 133, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Muriel, P. Role of Free Radicals in Liver Diseases. Hepatol. Int. 2009, 3, 526–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seitz, H.K.; Stickel, F. Risk Factors and Mechanisms of Hepatocarcinogenesis with Special Emphasis on Alcohol and Oxidative Stress. Biol. Chem. 2006, 387, 349–360. [Google Scholar] [CrossRef]
- Marx, J. Cancer research. Inflammation and Cancer: The Link Grows Stronger. Science 2004, 306, 966–968. [Google Scholar] [CrossRef] [PubMed]
- Chartampilas, E.; Rafailidis, V.; Georgopoulou, V.; Kalarakis, G.; Hatzidakis, A.; Prassopoulos, P. Current Imaging Diagnosis of Hepatocellular Carcinoma. Cancers 2022, 14, 3997. [Google Scholar] [CrossRef]
- Jiang, H.Y.; Chen, J.; Xia, C.C.; Cao, L.K.; Duan, T.; Song, B. Noninvasive Imaging of Hepatocellular Carcinoma: From Diagnosis to Prognosis. World J. Gastroenterol. 2018, 24, 2348–2362. [Google Scholar] [CrossRef] [PubMed]
- Kundu, K.; Knight, S.F.; Willett, N.; Lee, S.; Taylor, W.R.; Murthy, N. Hydrocyanines: A Class of Fluorescent Sensors That Can Image Reactive Oxygen Species in Cell Culture, Tissue, and in Vivo. Angew. Chem.—Int. Ed. 2009, 48, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, K.A.; Dorkin, J.R.; Vegas, A.J.; Chang, P.H.; Veiseh, O.; Matthews, J.; Fenton, O.S.; Zhang, Y.; Olejnik, K.T.; Yesilyurt, V.; et al. Degradable Lipid Nanoparticles with Predictable in Vivo SiRNA Delivery Activity. Nat. Commun. 2014, 5, 4277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zetasizer Nano User Manual (English); MAN0485; Malvern Instruments Ltd.: Malvern, UK, 2013.
- Quant-IT TM RiboGreen TM RNA Reagent and Kit; MAN0002073; Thermo Fisher Scientific Inc.: Eugene, OR, USA, 2022.
- Asghar, M.N.; Emani, R.; Alam, C.; Helenius, T.O.; Grönroos, T.J.; Sareila, O.; Din, M.U.; Holmdahl, R.; Hänninen, A.; Toivola, D.M. In Vivo Imaging of Reactive Oxygen and Nitrogen Species in Murine Colitis. Inflamm. Bowel Dis. 2014, 20, 1435–1447. [Google Scholar] [CrossRef] [PubMed]
- Fan, N.; Silverman, S.M.; Liu, Y.; Wang, X.; Kim, B.J.; Tang, L.; Clark, A.F.; Liu, X.; Pang, I.H. Rapid Repeatable in Vivo Detection of Retinal Reactive Oxygen Species. Exp. Eye Res. 2017, 161, 71–81. [Google Scholar] [CrossRef]
- Belousov, V.V.; Fradkov, A.F.; Lukyanov, K.A.; Staroverov, D.B.; Shakhbazov, K.S.; Terskikh, A.V.; Lukyanov, S. Genetically Encoded Fluorescent Indicator for Intracellular Hydrogen Peroxide. Nat. Methods 2006, 3, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Sergeeva, O.; Abakumova, T.; Kurochkin, I.; Ialchina, R.; Kosyreva, A.; Prikazchikova, T.; Varlamova, V.; Shcherbinina, E.; Zatsepin, T. Level of Murine Ddx3 Rna Helicase Determines Phenotype Changes of Hepatocytes In Vitro and In Vivo. Int. J. Mol. Sci. 2021, 22, 6958. [Google Scholar] [CrossRef]
- Leboeuf, D.; Abakumova, T.; Prikazchikova, T.; Rhym, L.; Anderson, D.G.; Zatsepin, T.S.; Piatkov, K.I. Downregulation of the Arg/N-Degron Pathway Sensitizes Cancer Cells to Chemotherapy In Vivo. Mol. Ther. 2020, 28, 1092–1104. [Google Scholar] [CrossRef] [PubMed]
- Agita, A.; Thaha, M. Inflammation, Immunity, and Hypertension. Acta Med. Indones. 2017, 49, 158–165. [Google Scholar]
- Sánchez-Valle, V.; Chávez-Tapia, N.C.; Uribe, M.; Méndez-Sánchez, N. Role of Oxidative Stress and Molecular Changes in Liver Fibrosis: A Review. Curr. Med. Chem. 2012, 19, 4850–4860. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Ishihara, H. Difference in the Lipid Nanoparticle Technology Employed in Three Approved SiRNA (Patisiran) and MRNA (COVID-19 Vaccine) Drugs. Drug Metab Pharm. 2021, 41, 100424. [Google Scholar] [CrossRef] [PubMed]
- Akinc, A.; Maier, M.A.; Manoharan, M.; Fitzgerald, K.; Jayaraman, M.; Barros, S.; Ansell, S.; Du, X.; Hope, M.J.; Madden, T.D.; et al. The Onpattro Story and the Clinical Translation of Nanomedicines Containing Nucleic Acid-Based Drugs. Nat. Nanotechnol. 2019, 14, 1084–1087. [Google Scholar] [CrossRef]
- Blakney, A.K.; McKay, P.F.; Hu, K.; Samnuan, K.; Jain, N.; Brown, A.; Thomas, A.; Rogers, P.; Polra, K.; Sallah, H.; et al. Polymeric and Lipid Nanoparticles for Delivery of Self-Amplifying RNA Vaccines. J. Control. Release 2021, 338, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Oberli, M.A.; Reichmuth, A.M.; Dorkin, J.R.; Mitchell, M.J.; Fenton, O.S.; Jaklenec, A.; Anderson, D.G.; Langer, R.; Blankschtein, D. Lipid Nanoparticle Assisted MRNA Delivery for Potent Cancer Immunotherapy. Nano. Lett. 2017, 17, 1326–1335. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarek, J.C.; Patel, A.K.; Kauffman, K.J.; Fenton, O.S.; Webber, M.J.; Heartlein, M.W.; DeRosa, F.; Anderson, D.G. Polymer–Lipid Nanoparticles for Systemic Delivery of MRNA to the Lungs. Angew. Chem.—Int. Ed. 2016, 55, 13808–13812. [Google Scholar] [CrossRef] [Green Version]
- Maeki, M.; Uno, S.; Niwa, A.; Okada, Y.; Tokeshi, M. Microfluidic Technologies and Devices for Lipid Nanoparticle-Based RNA Delivery. J. Control. Release 2022, 344, 80–96. [Google Scholar] [CrossRef]
- Lopes, C.; Cristóvão, J.; Silvério, V.; Lino, P.R.; Fonte, P. Microfluidic Production of MRNA-Loaded Lipid Nanoparticles for Vaccine Applications. Expert Opin. Drug Deliv. 2022, 19, 1381–1395. [Google Scholar] [CrossRef]
- Zhang, R.; El-Mayta, R.; Murdoch, T.J.; Warzecha, C.C.; Billingsley, M.M.; Shepherd, S.J.; Gong, N.; Wang, L.; Wilson, J.M.; Lee, D.; et al. Helper Lipid Structure Influences Protein Adsorption and Delivery of Lipid Nanoparticles to Spleen and Liver. Biomater. Sci. 2021, 9, 1449–1463. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, J.A.; Myhre, J.L.; Chen, S.; Tam, Y.Y.C.; Danescu, A.; Richman, J.M.; Cullis, P.R. Design of Lipid Nanoparticles for in Vitro and in Vivo Delivery of Plasmid DNA. Nanomedicine 2017, 13, 1377–1387. [Google Scholar] [CrossRef]
- Danaei, M.; Dehghankhold, M.; Ataei, S.; Hasanzadeh Davarani, F.; Javanmard, R.; Dokhani, A.; Khorasani, S.; Mozafari, M.R. Impact of Particle Size and Polydispersity Index on the Clinical Applications of Lipidic Nanocarrier Systems. Pharmaceutics 2018, 10, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaradat, E.; Weaver, E.; Meziane, A.; Lamprou, D.A. Microfluidic Paclitaxel-Loaded Lipid Nanoparticle Formulations for Chemotherapy. Int. J. Pharm. 2022, 628, 122320. [Google Scholar] [CrossRef]
- Prakash, G.; Shokr, A.; Willemen, N.; Bashir, S.M.; Shin, S.R.; Hassan, S. Microfluidic Fabrication of Lipid Nanoparticles for the Delivery of Nucleic Acids. Adv. Drug Deliv. Rev. 2022, 184, 114197. [Google Scholar] [CrossRef] [PubMed]
- Pak, V.V.; Ezeriņa, D.; Lyublinskaya, O.G.; Pedre, B.; Tyurin-Kuzmin, P.A.; Mishina, N.M.; Thauvin, M.; Young, D.; Wahni, K.; Martínez Gache, S.A.; et al. Ultrasensitive Genetically Encoded Indicator for Hydrogen Peroxide Identifies Roles for the Oxidant in Cell Migration and Mitochondrial Function. Cell Metab. 2020, 31, 642–653. [Google Scholar] [CrossRef] [PubMed]
- Ermakova, Y.G.; Bilan, D.S.; Matlashov, M.E.; Mishina, N.M.; Markvicheva, K.N.; Subach, O.M.; Subach, F.V.; Bogeski, I.; Hoth, M.; Enikolopov, G.; et al. Red Fluorescent Genetically Encoded Indicator for Intracellular Hydrogen Peroxide. Nat. Commun. 2014, 5, 5222. [Google Scholar] [CrossRef] [Green Version]
- Godbout, K.; Tremblay, J.P. Delivery of RNAs to Specific Organs by Lipid Nanoparticles for Gene Therapy. Pharmaceutics 2022, 14, 2129. [Google Scholar] [CrossRef]
- Zamboni, C.G.; Kozielski, K.L.; Vaughan, H.J.; Nakata, M.M.; Kim, J.; Higgins, L.J.; Pomper, M.G.; Green, J.J. Polymeric Nanoparticles as Cancer-Specific DNA Delivery Vectors to Human Hepatocellular Carcinoma. J. Control. Release 2017, 263, 18–28. [Google Scholar] [CrossRef]
- Huang, B.K.; Ali, S.; Stein, K.T.; Sikes, H.D. Interpreting Heterogeneity in Response of Cells Expressing a Fluorescent Hydrogen Peroxide Biosensor. Biophys. J. 2015, 109, 2148–2158. [Google Scholar] [CrossRef] [Green Version]
- Prunty, M.C.; Aung, M.H.; Hanif, A.M.; Allen, R.S.; Chrenek, M.A.; Boatright, J.H.; Thule, P.M.; Kundu, K.; Murthy, N.; Pardue, M.T. In Vivo Imaging of Retinal Oxidative Stress Using a Reactive Oxygen Species-Activated Fluorescent Probe. Investig. Ophthalmol. Vis. Sci. 2015, 56, 5862–5870. [Google Scholar] [CrossRef] [Green Version]




| Formulation | Cationic Lipid | Particle Size, nm | PDI | DNA Concentration in the Particles, ng/µL | DNA Loading Efficiency, % |
|---|---|---|---|---|---|
| Manual | DOPE | 177.7 ± 48.1 | 0.18 | 6.3 ± 1.1 | 10.8 ± 1.9% |
| Manual | DOPC | 214.3±15 | 0.2 | 8 | 13.6% |
| Microfluidic | DOPE | 120 | 0.2 | 34 | 62% |
| Microfluidic | DOPC | 108 ± 12 | 0.22 ± 0.02 | 36 ± 5 | 64 ± 7% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shashkovskaya, V.S.; Vetosheva, P.I.; Shokhina, A.G.; Aparin, I.O.; Prikazchikova, T.A.; Mikaelyan, A.S.; Kotelevtsev, Y.V.; Belousov, V.V.; Zatsepin, T.S.; Abakumova, T.O. Delivery of Lipid Nanoparticles with ROS Probes for Improved Visualization of Hepatocellular Carcinoma. Biomedicines 2023, 11, 1783. https://doi.org/10.3390/biomedicines11071783
Shashkovskaya VS, Vetosheva PI, Shokhina AG, Aparin IO, Prikazchikova TA, Mikaelyan AS, Kotelevtsev YV, Belousov VV, Zatsepin TS, Abakumova TO. Delivery of Lipid Nanoparticles with ROS Probes for Improved Visualization of Hepatocellular Carcinoma. Biomedicines. 2023; 11(7):1783. https://doi.org/10.3390/biomedicines11071783
Chicago/Turabian StyleShashkovskaya, Vera S., Polina I. Vetosheva, Arina G. Shokhina, Ilya O. Aparin, Tatiana A. Prikazchikova, Arsen S. Mikaelyan, Yuri V. Kotelevtsev, Vsevolod V. Belousov, Timofei S. Zatsepin, and Tatiana O. Abakumova. 2023. "Delivery of Lipid Nanoparticles with ROS Probes for Improved Visualization of Hepatocellular Carcinoma" Biomedicines 11, no. 7: 1783. https://doi.org/10.3390/biomedicines11071783
APA StyleShashkovskaya, V. S., Vetosheva, P. I., Shokhina, A. G., Aparin, I. O., Prikazchikova, T. A., Mikaelyan, A. S., Kotelevtsev, Y. V., Belousov, V. V., Zatsepin, T. S., & Abakumova, T. O. (2023). Delivery of Lipid Nanoparticles with ROS Probes for Improved Visualization of Hepatocellular Carcinoma. Biomedicines, 11(7), 1783. https://doi.org/10.3390/biomedicines11071783