Cyclic Production of Galacto-Oligosaccharides through Ultrafiltration-Assisted Enzyme Recovery
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Enzyme Activity Assay
2.3. Enzymatic Conversion in Stirred Tank Reactor (STR)
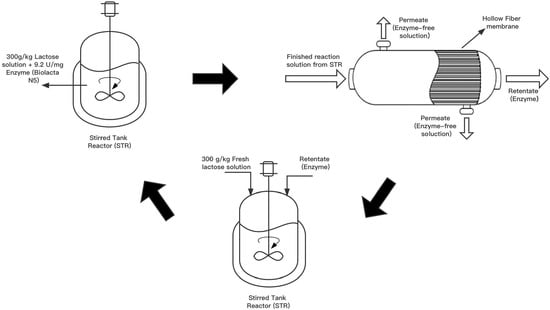
2.4. Cyclic Production of GOS in an Ultrafiltration-Assisted Reactor (Cyclic-EMR)
- In the first step, a traditional STR was employed to carry out a batchwise reaction. A 9.5 kg reaction solution with an initial lactose concentration of 300 g·kg−1 was introduced in the reactor TK-1. The reaction was performed at 50 °C and pH 6.0, using an initial enzyme activity of 8307 U·kg−1. Samples were regularly taken from the reactor and heat-treated at 90 °C for 30 min prior to the saccharides analysis by HPLC.
- In the second step, the membrane unit M-1 was attached to the reactor, and the reaction liquid was filtered through UF in an inside-out flow configuration at 0.5 bar transmembrane pressure until 8.4 kg of permeate was collected.
- In the third step, the membrane module M-1 was de-attached from the plant. A total of 8.4 kg of fresh substrate solution consisting of 300 g·kg−1 of lactose was added into the concentrated enzyme solution in the reactor. Then, step 1 of the next cycle was started. The de-attached membrane was subject to a cleaning procedure, as detailed in Section 2.5.
2.5. Membrane Regeneration Procedure
2.6. Analysis of Progress Curves
2.7. High Performance Liquid Chromatography (HPLC) Analysis
3. Results
3.1. STR Performance
3.2. Cyclic-EMR Performance
3.3. Quantification of Enzyme Losses
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
List of Symbols
Abbreviations
References
- Galacto-oligosaccharide Market Segmentation By End User (Food & Beverage Industry, Pharmaceutical, Personal Care and Animal Feed)–Global Demand Analysis & Opportunity Outlook 2028. Available online: https://www.researchnester.com/reports/galacto-oligosaccharide-market/2814 (accessed on 1 February 2022).
- Illanes, A.; Vera, C. Enzymatic production of galacto-oligosaccharides. In Lactose-Derived Prebiotics: A Process Perspective; Elsevier Inc.: Amsterdam, The Netherlands, 2016; pp. 111–189. [Google Scholar]
- Pázmándi, M.; Maráz, A.; Ladányi, M.; Kovács, Z. The impact of membrane pretreatment on the enzymatic production of whey-derived galacto-oligosaccharides. J. Food Process. Eng. 2018, 41, e12649. [Google Scholar] [CrossRef]
- Pires, A.F.; Marnotes, N.G.; Rubio, O.D.; Garcia, A.C.; Pereira, C.D. Dairy by-products: A review on the valorization of whey and second cheese whey. Foods 2021, 10, 1067. [Google Scholar] [CrossRef]
- Osman, A.; Tzortzis, G.; Rastall, R.A.; Charalampopoulos, D. A comprehensive investigation of the synthesis of prebiotic galactooligosaccharides by whole cells of Bifidobacterium bifidum NCIMB 41171. J. Biotechnol. 2010, 150, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; O’sullivan, D. Production of galactooligosaccharides using a hyperthermophilic β-galactosidase in permeabilized whole cells of Lactococcus lactis. J. Dairy Sci. 2014, 97, 694–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, T.; Pázmándi, M.; Galambos, I.; Kovács, Z. Continuous production of galacto-oligosaccharides by an enzyme membrane reactor utilizing free enzymes. Membranes 2020, 10, 203. [Google Scholar] [CrossRef] [PubMed]
- Das, R.; Sen, D.; Sarkar, A.; Bhattacharyya, S.; Bhattacharjee, C. A comparative study on the production of galacto-oligosaccharide from whey permeate in recycle membrane reactor and in enzymatic batch reactor. Ind. Eng. Chem. Res. 2011, 50, 806–816. [Google Scholar] [CrossRef]
- Hackenhaar, C.R.; Spolidoro, L.S.; Flores, E.E.E.; Klein, M.P.; Hertz, P.F. Batch synthesis of galactooligosaccharides from co-products of milk processing using immobilized β-galactosidase from Bacillus circulans. Biocatal. Agric. Biotechnol. 2021, 36, 102136. [Google Scholar] [CrossRef]
- Huerta, L.M.; Vera, C.; Guerrero, C.; Wilson, L.; Illanes, A. Synthesis of galacto-oligosaccharides at very high lactose concentrations with immobilized β-galactosidases from Aspergillus oryzae. Process. Biochem. 2011, 46, 245–252. [Google Scholar] [CrossRef]
- Pázmándi, M.; Kovács, Z.; Balga, E.; Kovács, M.; Maráz, A. Production of high-purity galacto-oligosaccharides by depleting glucose and lactose from galacto-oligosaccharide syrup with yeasts. Yeast 2020, 37, 515–530. [Google Scholar] [CrossRef]
- Gänzle, M.G. Enzymatic synthesis of galacto-oligosaccharides and other lactose derivatives (hetero-oligosaccharides) from lactose. Int. Dairy J. 2012, 22, 116–122. [Google Scholar] [CrossRef]
- Fischer, C.; Kleinschmidt, T. Synthesis of galactooligosaccharides in milk and whey: A review. Compr. Rev. Food Sci. Food Saf. 2018, 17, 678–697. [Google Scholar] [CrossRef] [PubMed]
- Füreder, V.; Rodriguez-Colinas, B.; Cervantes, F.V.; Fernandez-Arrojo, L.; Poveda, A.; Jimenez-Barbero, J.; Ballesteros, A.O.; Plou, F.J. Selective synthesis of galactooligosaccharides containing β (1 → 3) linkages with β-galactosidase from Bifidobacterium bifidum (Saphera). J. Agric. Food Chem. 2020, 68, 4930–4938. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhu, J.; Liu, L.; Yaqoob, M.U.; Pei, X.; Tao, W.; Xiao, Z.; Sun, W.; Wang, M. Optimization for galactooligosaccharides synthesis: A potential alternative for gut health and immunity. Life Sci. 2020, 245, 117353. [Google Scholar] [CrossRef] [PubMed]
- Scott, F.; Vera, C.; Conejeros, R. Technical and economic analysis of industrial production of lactose-derived prebiotics with focus on galacto-oligosaccharides. In Lactose-Derived Prebiotics: A Process Perspective; Elsevier Inc.: Amsterdam, The Netherlands, 2016; pp. 261–284. [Google Scholar]
- Albayrak, N.; Yang, S.T. Production of galacto-oligosaccharides from lactose by Aspergillus oryzae β-galactosidase immobilized on cotton cloth. Biotechnol. Bioeng. 2002, 77, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Torres, P.; Batista-Viera, F. Improved biocatalysts based on Bacillus circulans β-galactosidase immobilized onto epoxy-activated acrylic supports: Applications in whey processing. J. Mol. Catal. B: Enzym. 2012, 83, 57–64. [Google Scholar] [CrossRef]
- Warmerdam, A.; Zisopoulos, F.K.; Boom, R.M.; Janssen, A.E. Kinetic characterization of galacto-oligosaccharide (GOS) synthesis by three commercially important β-galactosidases. Biotechnol. Prog. 2014, 30, 38–47. [Google Scholar] [CrossRef]
- Fujimoto, H.; Miyasato, M.; Ito, Y.; Sasaki, T.; Ajisaka, K. Purification and properties of recombinant β-galactosidase from Bacillus circulans. Glycoconj. J. 1998, 15, 155–160. [Google Scholar] [CrossRef]
- Huang, J.; Zhu, S.; Zhao, L.; Chen, L.; Du, M.; Zhang, C.; Yang, S.-T. A novel β-galactosidase from Klebsiella oxytoca ZJUH1705 for efficient production of galacto-oligosaccharides from lactose. Appl. Microbiol. Biotechnol. 2020, 104, 6161–6172. [Google Scholar] [CrossRef]
- Urrutia, P.; Mateo, C.; Guisán, J.M.; Wilson, L.; Illanes, A. Immobilization of Bacillus circulans β-galactosidase and its application in the synthesis of galacto-oligosaccharides under repeated-batch operation. Biochem. Eng. J. 2013, 77, 41–48. [Google Scholar] [CrossRef]
- An, S.M.; Wu, J.H.; Qian, L.F.; Gao, Y.L.; Wu, Y.; Yu, G.P. Applications of Ultrafiltration-nanofiltration Membrane Continuous Combination Technology for Refining of Milk-Derived Oligosaccharides. In Advanced Materials Research; Trans Tech Publications Ltd.: Bäch, Switzerland, 2013; pp. 1429–1434. [Google Scholar]
- Matella, N.; Dolan, K.; Lee, Y. Comparison of galactooligosaccharide production in free-enzyme ultrafiltration and in immobilized-enzyme systems. J. Food Sci. 2006, 71, C363–C368. [Google Scholar] [CrossRef]
- Warmerdam, A.; Boom, R.M.; Janssen, A.E. β-galactosidase stability at high substrate concentrations. Springerplus 2013, 2, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Su, Z.; Luo, J.; Li, X.; Pinelo, M. Enzyme membrane reactors for production of oligosaccharides: A review on the interdependence between enzyme reaction and membrane separation. Sep. Purif. Technol. 2020, 243, 116840. [Google Scholar] [CrossRef]
- Córdova, A.; Astudillo, C.; Vera, C.; Guerrero, C.; Illanes, A. Performance of an ultrafiltration membrane bioreactor (UF-MBR) as a processing strategy for the synthesis of galacto-oligosaccharides at high substrate concentrations. J. Biotechnol. 2016, 223, 26–35. [Google Scholar] [CrossRef]
- Splechtna, B.; Nguyen, T.-H.; Haltrich, D. Comparison between discontinuous and continuous lactose conversion processes for the production of prebiotic galacto-oligosaccharides using β-galactosidase from Lactobacillus reuteri. J. Agric. Food Chem. 2007, 55, 6772–6777. [Google Scholar] [CrossRef] [PubMed]
- IBM SPSS Statistics for Windows; Version 27.0.; IBM Corp.: Armonk, NY, USA, 2020.
- Rodriguez-Colinas, B.; Poveda, A.; Jimenez-Barbero, J.; Ballesteros, A.O.; Plou, F.J. Galacto-oligosaccharide synthesis from lactose solution or skim milk using the β-galactosidase from Bacillus circulans. J. Agric. Food Chem. 2012, 60, 6391–6398. [Google Scholar] [CrossRef] [Green Version]
- Song, J.; Abe, K.; Imanaka, H.; Imamura, K.; Minoda, M.; Yamaguchi, S.; Nakanishi, K. Causes of the production of multiple forms of β-galactosidase by Bacillus circulans. Biosci. Biotechnol. Biochem. 2011, 75, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Vetere, A.; Paoletti, S. Separation and characterization of three β-galactosidases from Bacillus circulans. Biochim. Et Biophys. Acta Gen. Subj. 1998, 1380, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Boon, M.; Janssen, A.; Van der Padt, A. Modelling and parameter estimation of the enzymatic synthesis of oligosaccharides by β-galactosidase from Bacillus circulans. Biotechnol. Bioeng. 1999, 64, 558–567. [Google Scholar] [CrossRef]
- Palai, T.; Mitra, S.; Bhattacharya, P.K. Kinetics and design relation for enzymatic conversion of lactose into galacto-oligosaccharides using commercial grade β-galactosidase. J. Biosci. Bioeng. 2012, 114, 418–423. [Google Scholar] [CrossRef]
- Botelho, V.A.; Mateus, M.; Petrus, J.C.; de Pinho, M.N. Membrane Bioreactor for Simultaneous Synthesis and Fractionation of Oligosaccharides. Membranes 2022, 12, 171. [Google Scholar] [CrossRef]
- Córdova, A.; Astudillo, C.; Giorno, L.; Guerrero, C.; Conidi, C.; Illanes, A.; Cassano, A. Nanofiltration potential for the purification of highly concentrated enzymatically produced oligosaccharides. Food Bioprod. Process. 2016, 98, 50–61. [Google Scholar] [CrossRef]
- Córdova, A.; Astudillo, C.; Guerrero, C.; Vera, C.; Illanes, A. Assessment of the fouling mechanisms of an ultrafiltration membrane bioreactor during synthesis of galacto-oligosaccharides: Effect of the operational variables. Desalination 2016, 393, 79–89. [Google Scholar] [CrossRef]







| Enzyme Type/ Microorganism | Reactor Configuration | Reaction Conditions | Half-Life (h) | Deactivation Model | Activity Measurement Method | Ref. | ||
|---|---|---|---|---|---|---|---|---|
| Lactose Conc. | pH | Temp. (°C) | ||||||
| β-galactosidase/Aspergillus oryzae | Batch (FE) | 0 g·L−1 | 4.5 | 40 | 399 | Equation (6) | DM * | [17] |
| 50 | 49 | |||||||
| 60 | 2 | |||||||
| Continuous (IE) | 200 g·L−1 | 4.5 | 40 | 10,040 | ||||
| 50 | 1155 | |||||||
| 60 | 49 | |||||||
| β-galactosidase/Aspergillus oryzae | Batch (FE) | 0.1 g·L−1 | 4.5 | 50 | 42 | Equation (2) | oNPG | [10] |
| 55 | 7.4 | |||||||
| 60 | 0.8 | |||||||
| Batch (IE) | 2% w·v−1 | 4.5 | 50 | 163–166 | ||||
| 55 | 9.3–20.5 | |||||||
| 60 | 0.9–1.9 | |||||||
| Biolacta N5/Bacillus circulans | Batch (FE) | 30% w·w−1 | 6 | 25 | 29 | Equation (6) | oNPG | [25] |
| 40 | 29 | |||||||
| 60 | 16 | |||||||
| Biolacta N5/Bacillus circulans | Batch (FE) | 4.6% w·w−1 | 6 | 50 | 12 | Equation (5) | oNPG | [18] |
| Batch (IE) | 4.6% w·w−1 | 6 | 50 | 21–387 | ||||
| Biolactasa-NTL CONC X2/Bacillus circulans | Batch (FE) | 0 g·L−1 | 6 | 60 | ~0.05 | Equation (3) | oNPG | [22] |
| Batch (IE) | 0 g·L−1 | 6 | 60 | 0.25–3 | ||||
| β-galactosidase/Klebsiella oxytoca ZJUH1705 | Batch (FE) β-gal 1 | 40% w·w−1 | 7 | 30 | 141.67 | Equation (6) | oNPG | [21] |
| 40 | 18.33 | |||||||
| 50 | 0.04 | |||||||
| 60 | 0.01 | |||||||
| Batch (FE) β-gal 2 | 40% w·w−1 | 7 | 30 | 88.33 | ||||
| 40 | 1.17 | |||||||
| 50 | 0.17 | |||||||
| 60 | 0.01 | |||||||
| Enzyme Activity (U·kg−1) | Parameters | Estimate | Std. Error | 95% Confidence Interval | F (2; df2) | p1 × p2 | ||
|---|---|---|---|---|---|---|---|---|
| Lower Bound | Upper Bound | |||||||
| 923 | −162.07 * | 3.11 | 155.30 | 168.84 | 28,504.6 * df2 = 12 | >0.99 * | −22.69 | |
| 0.14 * | 0.01 | 0.13 | 0.15 | |||||
| 2307 | −172.91 * | 2.92 | 166.56 | 179.27 | 5352.4 * df2 = 12 | 0.99 * | −70.03 | |
| 0.41 * | 0.03 | 0.35 | 0.46 | |||||
| 4615 | −180.19 * | 3.35 | 172.60 | 187.76 | 4539.8 * df2 = 9 | 0.99 * | −134.24 | |
| 0.75 * | 0.06 | 0.62 | 0.87 | |||||
| 6923 | −177.63 * | 1.64 | 174.07 | 181.20 | 6763.6 * df2 = 12 | 0.99 * | −220.44 | |
| 1.24 * | 0.08 | 1.08 | 1.41 | |||||
| 8307 | −179.05 * | 1.72 | 175.53 | 182.56 | 8630.4 * df2 = 29 | 0.99 * | −237.95 | |
| 1.33 * | 0.07 | 1.20 | 1.46 | |||||
| Saccharides Compounds | No-Intercept Linear Regression Slopes | R2 |
|---|---|---|
| DP2 | 0.03 * | 0.997 * |
| DP3 | 0.025 * | 0.991 * |
| DP4 | 0.004 * | 0.995 * |
| DP3-6 | 0.024 * | 0.997 * |
| Glucose | 0.007 * | 0.999 * |
| Cycles | |||||
|---|---|---|---|---|---|
| Fractions | No.1 | No.2 | No.3 | No.4 | No.5 |
| DP2 | 7077 | 2124 | 846 | 388 | 329 |
| DP3 | 7925 | 2480 | 890 | 379 | 306 |
| DP3-6 | 6999 | 2081 | 785 | 355 | 295 |
| Glucose | 7903 | 2588 | 992 | 448 | 400 |
| Mean | 7476 | 2318 | 878 | 392 | 333 |
| STDEV | 507 | 254 | 87 | 40 | 47 |
| Model | k1 | k2 | α1 | α2 | R2 | SSR |
|---|---|---|---|---|---|---|
| Equation (2) | 9.692 × 10−1 | 4.839 × 10−2 | 8.125 × 10−1 | 2.977 × 10−2 | 0.9974 | 7.338 × 10−3 |
| Equation (3) | 5.947 × 10−2 | 9.980 × 10−3 | 7.493 × 10−2 | 0 | 0.9987 | 3.535 × 10−3 |
| Equation (5) | 5.537 × 10−2 | 0 | 3.651 × 10−2 | 0 | 0.9986 | 3.674 × 10−3 |
| Equation (6) | 4.891 × 10−2 | 0 | 0 | 0 | 0.9957 | 1.194 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, T.; Kovács, Z.; Ladányi, M. Cyclic Production of Galacto-Oligosaccharides through Ultrafiltration-Assisted Enzyme Recovery. Processes 2023, 11, 225. https://doi.org/10.3390/pr11010225
Cao T, Kovács Z, Ladányi M. Cyclic Production of Galacto-Oligosaccharides through Ultrafiltration-Assisted Enzyme Recovery. Processes. 2023; 11(1):225. https://doi.org/10.3390/pr11010225
Chicago/Turabian StyleCao, Teng, Zoltán Kovács, and Márta Ladányi. 2023. "Cyclic Production of Galacto-Oligosaccharides through Ultrafiltration-Assisted Enzyme Recovery" Processes 11, no. 1: 225. https://doi.org/10.3390/pr11010225
APA StyleCao, T., Kovács, Z., & Ladányi, M. (2023). Cyclic Production of Galacto-Oligosaccharides through Ultrafiltration-Assisted Enzyme Recovery. Processes, 11(1), 225. https://doi.org/10.3390/pr11010225