Ability of Trichoderma hamatum Isolated from Plastics-Polluted Environments to Attack Petroleum-Based, Synthetic Polymer Films
Abstract
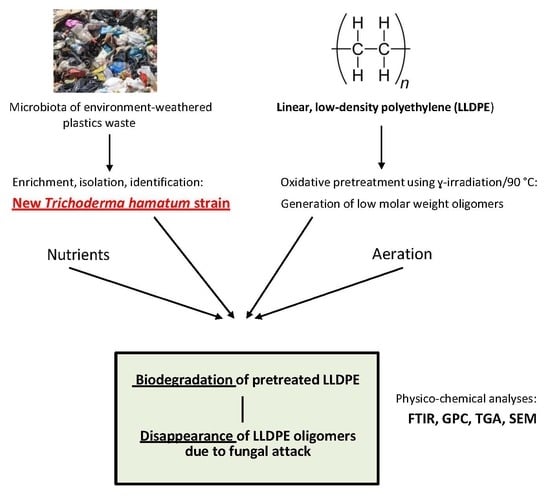
:1. Introduction
2. Materials and Methods
2.1. Isolation and Taxonomical Identification of Bacteria and Fungi from Plastic-Polluted Environments
2.2. Plastic Polymers and Their Pretreatment
2.3. Biodegradation Tests
2.4. Physicochemical Analyses
2.4.1. FTIR Analysis
2.4.2. TGA Analysis
2.4.3. GPC Analysis
2.4.4. Scanning Electron Microscopy (SEM)
3. Results and Discussion
3.1. Isolation and Identification of Microorganisms
3.2. Biodegradation of Polymer Plastic Films Measured by Gravimetric Analysis
3.3. Biodegradation of Polymer Plastic Films by T. Hamatum Measured by Physicochemical Methods
3.3.1. FTIR Analysis
3.3.2. GPC Analysis
3.3.3. TGA Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kale, S.K.; Deshmukh, A.G.; Dudhare, M.S.; Patil, V.B. Microbial degradation of plastic: A review. J. Biochem. Tech. 2015, 6, 952–961. [Google Scholar]
- Sivan, A. New perspectives in plastic biodegradation. Curr. Opin. Biotechnol. 2011, 22, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, T.; Shahid, M.; Azeem, F.; Rasul, I.; Shah, A.A.; Noman, M.; Hameed, A.; Manzoor, N.; Manzoor, I.; Muhammad, S. Biodegradation of plastics: Current scenario and future prospects for environmental safety. Environ. Sci. Pollut. Res. 2018, 25, 7287–7298. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.I.; Ahmed, S.; Robson, G.; Javed, I.; Ali, N.; Atiq, N.; Hameed, A. Isolation and molecular characterization of polyvinyl chloride (PVC) plastic degrading fungal isolates. J. Basic Microbiol. 2014, 54, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Geyer, R.; Jambeck, J.R.; Law, K.L. Production, use, and fate of all plastics ever made. Sci. Adv. 2017, 3, e1700782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Briassoulis, D.; Babou, E.; Hiskakis, M.; Kyrikou, I. Degradation in soil behavior of artificially aged polyethylene films with pro-oxidants. J. Appl. Polym. Sci. 2015, 132, 42289. [Google Scholar] [CrossRef]
- Fontanella, S.; Bonhomme, S.; Koutny, M.; Husarova, L.; Brusson, J.M.; Courdavault, J.P.; Pitteri, S.; Samuel, G.; Pichon, G.; Lemaire, J.; et al. Comparison of the biodegradability of various polyethylene films containing pro-oxidant additives. Polym. Degrad. Stab. 2010, 95, 1011–1021. [Google Scholar] [CrossRef]
- Restrepo-Flórez, J.M.; Bassi, A.; Thompson, M.R. Microbial degradation and deterioration of polyethylene—A review. Int. Biodeter. Biodegr. 2014, 88, 83–90. [Google Scholar] [CrossRef]
- Watanabe, T.; Ohtake, Y.; Asabe, H.; Murakami, N.; Furukawa, M. Biodegradability and degrading microbes of low-density polyethylene. J. Appl. Polym. Sci. 2009, 111, 551–559. [Google Scholar] [CrossRef]
- Das, M.P.; Kumar, S. Microbial deterioration of low density polyethylene by Aspergillus and Fusarium sp. Int. J. Chem. Tech. Res. 2014, 6, 299–305. [Google Scholar]
- Sindujaa, P.; Padmapriya, M.; Pramila, R.; Vijaya Ramesh, K. Bio-degradation of low density polyethylene (LDPE) by fungi isolated from marine water. Res. J. Biol. Sci. 2011, 6, 141–145. [Google Scholar]
- Singh, J.; Gupta, K.C. Screening and identification of low density polyethylene (LDPE) degrading soil fungi isolated from polythene polluted sites around Gwalior city (M.P.). Int. J. Curr. Microbiol. App. Sci. 2014, 3, 443–448. [Google Scholar]
- Singh, V.; Dubey, M.; Bhadauria, S. Microbial degradation of polyethylene (low density) by Aspergillus fumigatus and Penicillium sp. Asian J. Exp. Biol. Sci. 2012, 3, 498–503. [Google Scholar]
- Nowak, B.; Pajak, J.; Drozd-Bratkowicz, M.; Rymarz, G. Microorganisms participating in the biodegradation of modified polyethylene films in different soils under laboratory conditions. Int. Biodeter. Biodegr. 2011, 65, 757–767. [Google Scholar] [CrossRef]
- Czaczyk, K.; Bialas, W.; Myszka, K. Cell surface hydrophobicity of Bacillus spp. as a function of nutrient supply and lipopeptides biosynthesis and its role in adhesion. Pol. J. Microbiol. 2008, 57, 313–319. [Google Scholar]
- Motta, O.; Proto, A.; De Carlo, F.; De Caro, F.; Santoro, E.; Brunetti, L.; Capunzo, M. Utilization of chemically oxidized polystyrene as co-substrate by filamentous fungi. Int. J. Hyg. Environ. Health 2009, 212, 61–66. [Google Scholar] [CrossRef]
- Weiland, M.; Daro, A.; David, C. Biodegradation of thermally oxidized polyethylene. Polym. Degrad. Stab. 1995, 48, 275–289. [Google Scholar] [CrossRef]
- Arutchelvi, J.; Sudhakar, M.; Arkatkar, A.; Doble, M.; Bhaduri, S.; Uppara, P.V. Biodegradation of polyethylene and polypropylene. Indian J. Biotechnol. 2008, 7, 9–22. [Google Scholar]
- Iiyoshi, Y.; Tsutsumi, Y.; Nishida, T. Polyethylene degradation by lignin-degrading fungi and manganese peroxidase. J. Wood Sci. 1998, 44, 222–229. [Google Scholar] [CrossRef]
- Santo, M.; Weitsman, R.; Sivan, A. The role of the copper-binding enzyme—Laccase—In the biodegradation of polyethylene by the actinomycete Rhodococcus ruber. Int. Biodeter. Biodegr. 2013, 84, 204–210. [Google Scholar] [CrossRef]
- Khan, I.; Ray Dutta, J.; Ganesan, R. Lactobacillus sps. lipase mediated poly (ε-caprolactone) degradation. Int. J. Biol. Macromol. 2017, 95, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Kim, E.Y.; Yoo, Y.T.; Im, S.S. Effect of hydrophilicity on the biodegradability of polyesteramides. J. Appl. Polym. Sci. 2003, 90, 2708–2714. [Google Scholar] [CrossRef]
- Cerdà-Cuéllar, M.; Kint, D.P.R.; Muñoz-Guerra, S.; Marqués-Calvo, M.S. Biodegradability of aromatic building blocks for poly (ethylene terephthalate) copolyesters. Polym. Degrad. Stab. 2004, 85, 865–871. [Google Scholar] [CrossRef]
- Kleeberg, I.; Welzel, K.; VandenHeuvel, J.; Müller, R.J.; Deckwer, W.D. Characterization of a new extracellular hydrolase from Thermobifida fusca degrading aliphatic-aromatic copolyesters. Biomacromolecules 2005, 6, 262–270. [Google Scholar] [CrossRef]
- Corti, A.; Muniyasamy, S.; Vitali, M.; Imam, S.H.; Chiellini, E. Oxidation and biodegradation of polyethylene films containing pro-oxidant additives: Synergistic effects of sunlight exposure, thermal aging and fungal biodegradation. Polym. Degrad. Stab. 2010, 95, 1106–1114. [Google Scholar] [CrossRef]
- Tourova, T.P.; Sokolova, D.S.; Nazina, T.N.; Gruzdev, D.S.; Laptev, A.B. Phylogenetic diversity of microbial communities from the surface of polyethylene terephthalate materials exposed to different water environments. Microbiology 2020, 89, 96–106. [Google Scholar] [CrossRef]
- Tien, M.; Kirk, T.K. Lignin peroxidase of Phanerochaete chrysosporium. Method. Enzymol. 1988, 161, 238–249. [Google Scholar]
- Mabrouk, A.M.; Kheiralla, Z.H.; Hamed, E.R.; Youssry, A.A.; Abd, A.A.A. Screening of some marine-derived fungal isolates for lignin degrading enzymes (LDEs) production. Agric. Biol. J. N. Am. 2010, 1, 591–599. [Google Scholar]
- Archibald, F.S. A new assay for lignin-type peroxidases employing the dye Azure, B. Appl. Environ. Microbiol. 1992, 58, 3110–3116. [Google Scholar] [CrossRef] [Green Version]
- Kelley, J.; Yaghmaie, P.A. Screening of fungal strains employed in the testing of plastics materials. Int. Biodeterior. Biodegr. 2001, 48, 84–93. [Google Scholar] [CrossRef]
- Novotný, Č.; Malachová, K.; Adamus, G.; Kwiecień, M.; Lotti, N.; Soccio, M.; Verney, V.; Fava, F. Deterioration of irradiation/high-temperature pretreated, linear low-density polyethylene (LLDPE) by Bacillus amyloliquefaciens. Int. Biodeter. Biodegrad. 2018, 132, 259–267. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Wani, A.A.; Surakasi, V.P.; Siddharth, J.; Raghavan, R.G.; Patole, M.S.; Ranade, D.; Shouche, Y.S. Molecular analyses of microbial diversity associated with the Lonar soda lake in India: An impact crater in a basalt area. Res. Microbiol. 2006, 157, 928–937. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Mezei, L.M.; Storts, D.R. Purification of PCR Products in PCR Technology: Current Innovations; Griffin, H.G., Griffin, A.M., Eds.; CRC Press: Boca Raton, FL, USA, 1994; p. 21. [Google Scholar]
- Bochner, B. “Breathprints” at the microbial level. ASM News 1989, 55, 536–539. [Google Scholar]
- Lee, B.; Pometto, A.L.; Fratzke, A.; Bailey, T.B. Biodegradation of degradable plastic polyethylene by Phanerochaete and Streptomyces species. Appl. Environ. Microbiol. 1991, 57, 678–685. [Google Scholar] [CrossRef] [Green Version]
- Sheik, S.; Chandrashekar, K.R.; Swaroop, K.; Somashekarappa, H.M. Biodegradation of gamma irradiated low density polyethylene and polypropylene by endophytic fungi. Int. Biodeter. Biodegr. 2015, 105, 21–29. [Google Scholar] [CrossRef]
- Brunner, I.; Fischer, M.; Rüthi, J.; Stierli, B.; Frey, B. Ability of fungi isolated from plastic debris floating in the shoreline of a lake to degrade plastics. PLoS ONE 2018, 13, e0202047. [Google Scholar] [CrossRef] [Green Version]
- Syranidou, E.; Karkanorachaki, K.; Amorotti, F.; Franchini, M.; Repouskou, E.; Kaliva, M.; Vamvakaki, M.; Kolvenbach, B.; Fava, F.; Corvini, P.F.-X.; et al. Biodegradation of weathered polystyrene films in seawater microcosms. Sci. Rep. 2017, 7, 17991. [Google Scholar] [CrossRef] [Green Version]
- European Commission DGXI.E.3. The Behaviour of PVC in Landfill, Final Report February 2000, ARGUS in Association with University Rostock-Prof. Spillmann, Carl Bro a|s and Sigma Plan, S.A. Available online: http://ec.europa.eu/environment/waste/studies/pvc/landfill.pdf (accessed on 4 January 2017).
- Liang, D.W.; Zhang, T.; Fang, H.H.P.; He, J. Phthalates biodegradation in the environment. Appl. Microbiol. Biotechnol. 2008, 80, 183–198. [Google Scholar] [CrossRef]
- Giacomucci, L.; Raddadi, N.; Soccio, M.; Lotti, N.; Fava, F. Polyvinyl chloride biodegradation by Pseudomonas citronellolis and Bacillus flexus. New Biotechnol. 2019, 52, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Mersiowski, I.; Weller, M.; Ejlertsson, J. Fate of plasticised PVC products under landfill conditions: A laboratory-scale landfill simulation reactor study. Water Res. 2001, 35, 3063–3070. [Google Scholar] [CrossRef]
- Reddy, M.M.; Deighton, M.; Gupta, R.K.; Bhattacharya, S.N.; Parthasarathy, R. Biodegradation of oxo-biodegradable polyethylene. J. Appl. Polym. Sci. 2009, 111, 1426–1432. [Google Scholar] [CrossRef]




| Environmental Origin | Strain Identity (GenBank Accession No.) | Q-Coverage, % (Error Value) |
|---|---|---|
| Compost (Grenoble, France) | Vibrio sp. (DQ146981.1) | 99 (0) |
| Clostridium roseum (KM999946.1) | 71 (0) | |
| Bacillus sp. (JX202600.1) | 100 (0) | |
| Bacillus sp. (KJ162135) | 100 (0) | |
| Mulching Film From Soil (Belgium) | Pseudomonas poe (JN897284) | 100 (0) |
| Plastics From Soil Along Highway (Belgium) | Delftia sp. (KF896097) | 100 (0) |
| Uncultured Klebsiella sp. (JN873189) | 100 (0) | |
| Compost (Schendelbeke, Belgium) | Achromobacter sp. (KP670417.1) | 100 (1.00 × 10−112) |
| Vibrio sp. (DQ146981.1) | 99 (0) | |
| Bacillus licheniformis (KM226937.1) | 97 (0) | |
| Bacillus amyloliquefaciens JB4 (KT185076) 1 | 100 (0) | |
| Bacillus amyloliquefaciens (KJ469792.1) | 100 (0) | |
| Bacillus amyloliquefaciens (KJ545589) | 100 (0) | |
| Bacillus amyloliquefaciens (CP006890) | 100 (0) | |
| Bacillus amyloliquefaciens (CP010556.1) | 99 (0) | |
| Bacillus pumilus (DQ275671.1) | 97 (0) | |
| Bacillus subtilis (KJ865584.1) | 100 (0) | |
| Bacterium ZI-9 (JQ342232.1) | 99 (0) | |
| Sludge From Anaerobic Digester (Treviso, Italy) | Bacillus sp. (CP009938.1) | 100 (0) |
| Bacillus sp. (EF582419) | 100 (0) | |
| Bacillus amyloliquefaciens (KJ545589) | 100 (0) | |
| Plastics From Landfill (Styron, Germany) | Bacillus cereus (KF805048) | 100 (3.00 × 10−164) |
| Bacillus sp. (FJ596521.1) | 100 (0) | |
| Bacillus subtilis (KJ604979.1) | 100 (0) | |
| Klebsiella oxytoca (CP003218) | 100 (0) | |
| Alcaligenaceae sp. (AB847924.1) | 99 (0) |
| Environmental Origin | 18 S Sequence | ITS Sequence | ||
|---|---|---|---|---|
| Strain Identity (GenBank Access. No.) | Q–Coverage, % (Error Value) | Strain Identity (GenBank Access. No.) | Q–Coverage. % (Error Value) | |
| Compost Sample (Grenoble, France) | - | - | Filobasidium floriforme (KF971359) | 100 (0) |
| Mulching Film from Soil (Belgium) | Fusarium oxysporum (KF562839) | 100 (0) | Fusarium oxysporum (JF776163) | 100 (0) |
| Mucor circinelloides (JQ014009) | 100 (0) | Mucor circinelloides (HQ285608) | 99 (0) | |
| Fusarium oxysporum (KF562839) | 100 (0) | Fusarium oxysporum (KC202938) | 100 (0) | |
| Trametes sp. (FJ515315) | 99 (0) | Uncultured Fungus (KF800596) | 100 (0) | |
| Plastics from Soil Along Highway (Belgium) | Uncultured fungus (AB534505) | 100 (0) | Fusarium sp. (JQ388248) | 100 (0) |
| Hypocrea muroiana (JN941682) | 100 (0) | Trichoderma hamatum HF4 1 (FR872741) | 100 (0) | |
| Uncultured fungus (GU306002) | 99 (0) | Thanatephorus cucumeris (FR670341) | 100 (0) | |
| Plastic Samples from Composting Plant (Belgium) | - | - | Galactomyces geotrichum (DQ683112) | 100 (0) |
| Trichaptum abietinum CA 1 (FJ768676) | 100 (0) | Uncultured fungus (JF721422) | 100 (0) | |
| Byssochlamys nivea FK1 1 (M83256.1) | 100 (0) | - | - | |
| Pseudallescheria sp. (FN666094.1) | 99 (0) | Pseudallescheria sp. (AY939802.1) | 99 (0) | |
| - | - | Pseudallescheria boydii (AY213683.1) | 99 (0) | |
| - | - | Trametes suaveolens (KE573015) | 99 (0) | |
| Trametes sp. (FJ51531) | 99 (0) | Trametes gibbosa (KC525203) | 100 (0) | |
| Pseudallescheria ellipsoidea (U43911) | 100 (0) | Scedosporium apiospermum (JN207446) | 100 (0) | |
| Graphium sp. (FJ176832) | 100 (0) | - | - | |
| Sludge (Treviso, Italy) | Byssochlamys nivea JM5 1 (GU733368.1) | 99 (0) | - | - |
| Plastic Polymers | Character of Polymers | Microorganism | Weight Loss (%) | Abiotic Control (%) |
|---|---|---|---|---|
| LLDPE | Virgin | Trichoderma hamatum HF4 | 2.2 ± 1.2 | 1.3 ± 0.8 |
| Pretreated ɣ/T90 1 | Trichoderma hamatum HF4 | 3.9 ± 0.5 | 0.3 ± 0.1 | |
| LDPE | Virgin | Trichoderma hamatum HF4 | 0.5 ± 0.4 | 0.2 ± 0.1 |
| Pretreated ɣ/T150 1 | Trichoderma hamatum HF4 | 0.9 ± 0.1 | 0.2 ± 0.1 | |
| Pretreated UV/T60 1 | Trichoderma hamatum HF4 | 1.3 ± 0.4 | 0.4 ± 0.1 | |
| PS | Virgin | Trichoderma hamatum HF4 | 0.9 ± 0.4 | 0.4 ± 0.2 |
| PVC | Virgin | Trichoderma hamatum HF4 | 20.0 ± 0.5 | 9.9 ± 2.9 |
| Byssochlamys nivea FK1 | 18.4 ± 0.7 | |||
| Trichaptum abietinum CA | 17.5 ± 0.7 | |||
| Byssochlamys nivea JM5 | 15.5 ± 0.9 |
| Polymer | Treatment Conditions | Mn (g·mol−1) | Mw (g·mol−1) | Mw/Mn |
|---|---|---|---|---|
| Virgin LLDPE | No exposure | 75,500 | 163,400 | 2.16 |
| Pretreated LLDPE | No exposure | 1900 | 32,200 | 16.9 |
| Pretreated LLDPE | Abiotic control | 2700 | 38,500 | 14.25 |
| Pretreated LLDPE | 60-day exposure | 2700 | 38,000 | 14.07 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malachová, K.; Novotný, Č.; Adamus, G.; Lotti, N.; Rybková, Z.; Soccio, M.; Šlosarčíková, P.; Verney, V.; Fava, F. Ability of Trichoderma hamatum Isolated from Plastics-Polluted Environments to Attack Petroleum-Based, Synthetic Polymer Films. Processes 2020, 8, 467. https://doi.org/10.3390/pr8040467
Malachová K, Novotný Č, Adamus G, Lotti N, Rybková Z, Soccio M, Šlosarčíková P, Verney V, Fava F. Ability of Trichoderma hamatum Isolated from Plastics-Polluted Environments to Attack Petroleum-Based, Synthetic Polymer Films. Processes. 2020; 8(4):467. https://doi.org/10.3390/pr8040467
Chicago/Turabian StyleMalachová, Kateřina, Čeněk Novotný, Grażyna Adamus, Nadia Lotti, Zuzana Rybková, Michelina Soccio, Pavlína Šlosarčíková, Vincent Verney, and Fabio Fava. 2020. "Ability of Trichoderma hamatum Isolated from Plastics-Polluted Environments to Attack Petroleum-Based, Synthetic Polymer Films" Processes 8, no. 4: 467. https://doi.org/10.3390/pr8040467
APA StyleMalachová, K., Novotný, Č., Adamus, G., Lotti, N., Rybková, Z., Soccio, M., Šlosarčíková, P., Verney, V., & Fava, F. (2020). Ability of Trichoderma hamatum Isolated from Plastics-Polluted Environments to Attack Petroleum-Based, Synthetic Polymer Films. Processes, 8(4), 467. https://doi.org/10.3390/pr8040467