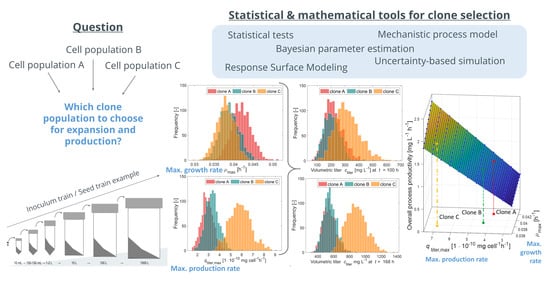
Considerations of the Impacts of Cell-Specific Growth and Production Rate on Clone Selection—A Simulation Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data from a Stability Study
2.2. Experimental Data and Set Up for Modeling Purposes
2.3. Cultivation Systems for Inoculum Train Simulations
2.4. Approximations of Empirical Growth Rates and Production Rates
2.5. Statistical Testing of the Differences in Means between Clonal Cell Populations
2.6. Mechanistic Model
2.7. Bayesian Parameter Estimation
2.8. Upstream Simulation—Software Tool
2.9. Uncertainty-Based Prediction
2.10. Response Surface Modeling
3. Results and Discussion
3.1. Analysis of Variabilities in Growth Rate and Production Rate for Five Clonal Cell Lines
- The averaged empirical growth rate of clone A, , is statistically significantly higher than the averaged empirical growth rate of clone B, , i.e., .
- The averaged empirical production rate of clone A, , is statistically significantly lower than averaged empirical production rate of clone B, , i.e., .
3.2. Model Adaption of a Mechanistic Cell Culture Model for Prediction Using Bayesian Parameter Estimation
3.2.1. Prior Knowledge
3.2.2. Posterior Distributions
3.3. Uncertainty-Based Upstream Process Simulation—Comparison of Three Clonal Populations with Different Growth and Production Rates
3.4. Impacts of Differences in Growth and Production Rates on Inoculum Train and Titer at Production Scale—General Considerations and a Decision Criterion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Shape parameter of a gamma distribution (-) | |
| Regression coefficients (impact) | |
| Scale parameter of a gamma distribution (-) | |
| Growth rate (h) | |
| Death rate (h) | |
| Maximum death rate (h) | |
| Minimum death rate (h) | |
| () | (Maximum) empirical cell-specific growth rate (h) |
| Maximum cell-specific growth rate (h) | |
| () | Averaged empirical growth rate of clone A (B) (h) |
| () | (Initial) ammonia concentration (mmol L) |
| () | (Initial) glucose concentration (mmol L) |
| () | (Initial) glutamine concentration (mmol L) |
| () | (Initial) lactate concentration (mmol L) |
| () | (Initial) limiting substrate concentration (mmol L) |
| () | (Initial) volumetric titer (product concentration) (mg L) |
| Volumetric titer (product concentration) at point in time (mg L) | |
| CHO | Chinese Hamster Ovary |
| CI | Confidence interval |
| cv | coefficient of variation |
| Difference in terms of growth rate (h) | |
| Difference in terms of production rate (mg cell h) | |
| Change of volume due to sampling [L h] | |
| i | Running index (-) |
| j | Running index (-) |
| Correction factor for ammonia uptake (-) | |
| Cell lysis constant (h) | |
| Monod kinetic constant for glucose (mmol L) | |
| Monod kinetic constant for glutamine (mmol L) | |
| Monod kinetic constant for limiting substrate (mmol L) | |
| Monod kinetic constant for glucose uptake (mmol L) | |
| Monod kinetic constant for glutamine uptake (mmol L) | |
| Monod kinetic constant for uptake of limiting substrate (mmol L) | |
| MAP | Maximum a posteriori |
| max | Maximum value |
| MCMC | Markov Chain Monte Carlo |
| () | (Maximum) cell-specific ammonia uptake rate (mmol cell h) |
| () | (Maximum) cell-specific glucose uptake rate (mmol cell h) |
| () | (Maximum) cell-specific glutamine uptake rate (mmol cell h) |
| () | (Maximum) cell-specific lactate uptake rate (mmol cell h) |
| () | (Max.) cell-specific uptake rate of limiting substrate (mmol cell h) |
| () | (Maximum) cell-specific production rate (mg cell h) |
| () | (Maximum) empirical cell-specific production rate (mg cell h) |
| , | Average empirical production rate of reference or compared clone |
| R | Coefficient of determination |
| RSM | Response surface models |
| sd | Standard deviation |
| SE | Standard error |
| t | Time (h) |
| Point in time with index i (h) | |
| V | Volume (L) |
| Viability (%) | |
| Total cell density (cells L) | |
| Viable cell density (cells L) | |
| Viable cell density at point in time with index i (cells L) | |
| Y | Arbitrary random variable (-) |
| Kinetic production constant for ammonia (mmol mmol) | |
| Kinetic production constant for lactate (mmol mmol) |
Appendix A
Appendix A.1. Choice of the Prior Distribution
Appendix B
Appendix B.1. Supplementary Figures









Appendix B.2. Supplementary Tables
| Variable/Parameter | Unit | Initial Value | Description |
|---|---|---|---|
| cells L | 5.3 | Total cell density | |
| cells L | 5.3 | Viable cell density | |
| mmol L | 5.9 | Glucose concentration | |
| mmol L | 3.3 | Glutamine concentration | |
| mmol L | 0.01 | Lactate concentration | |
| mmol L | 2.6 | Ammonia concentration | |
| mg L | 0 | Volumetric product (antibody) titer | |
| V | L | 0.01 | Volume |
| Model Parameter | Unit | Posterior Estimate/Fixed Value | Description |
| h | 0.042 | Maximum cell-specific growth rate | |
| mmol L | 0.03 | Monod kinetic constant for glucose | |
| mmol L | 0.03 | Monod kinetic constant for glutamine | |
| mmol L | 0.16 (fixed) | Monod kinetic constant for limiting substrate | |
| h | 1.0 | Minimum cell-specific death rate | |
| h | 0.08 | Maximum cell-specific death rate | |
| h | 1.2 | Cell lysis constant | |
| mmol cell h | 9.7 | Maximum cell-specific glucose uptake rate | |
| mmol L | 6.2 | Monod kinetic constant for glucose uptake | |
| mmol cell h | 1.1 | Maximum cell-specific glutamine uptake rate | |
| mmol L | 0.5 | Monod kinetic constant for glutamine uptake | |
| mmol cell h | 1.1 (fixed) | Max. cell-specific uptake rate of limiting substrate | |
| mmol L | 8.15 (fixed) | Monod kinetic constant for uptake of limiting substrate | |
| mmol mmol | 0.35 | Kinetic production constant for lactate | |
| mmol cell h | 1.0 (fixed) | Cell-specific maximum lactate uptake rate | |
| mmol mmol | 1.67 | Kinetic production constant for ammonia | |
| mmol cell h | 4.5 (fixed) | Cell-specific maximum ammonia uptake rate | |
| - | 1.9 (fixed) | Correction factor for ammonia uptake | |
| mg cell h | 3.9 | Cell-specific maximum production rate |
References
- Walsh, G. Biopharmaceutical benchmarks 2018. Nat. Biotechnol. 2018, 36, 1136–1145. [Google Scholar] [CrossRef] [PubMed]
- Wurm, F. CHO Quasispecies—Implications for Manufacturing Processes. Processes 2013, 1, 296. [Google Scholar] [CrossRef]
- Wurm, F.; Wurm, M. Cloning of CHO Cells, Productivity and Genetic Stability—A Discussion. Processes 2017, 5, 20. [Google Scholar] [CrossRef] [Green Version]
- Browne, S.; Al-Rubeai, M. Selection Methods for High-Producing Mammalian Cell Lines. Trends Biotechnol. 2007, 25, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Lakshmanan, M.; Kok, Y.J.; Lee, A.P.; Kyriakopoulos, S.; Lim, H.L.; Teo, G.; Poh, S.L.; Tang, W.Q.; Hong, J.; Tan, A.H.M.; et al. Multi–omics profiling of CHO parental hosts reveals cell line–specific variations in bioprocessing traits. Biotechnol. Bioeng. 2019, 116, 2117–2129. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, D.; Damjanovic, L.; Kaisermayer, C.; Sommeregger, W.; Gili, A.; Gasselhuber, B.; Castan, A.; Mayrhofer, P.; Grünwald-Gruber, C.; Kunert, R. Bioprocessing of Recombinant CHO-K1, CHO-DG44, and CHO-S: CHO Expression Hosts Favor Either mAb Production or Biomass Synthesis. Biotechnol. J. 2019, 14, e1700686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beketova, E.V.; Ibneeva, L.R.; Abdulina, Y.A.; Dergousova, E.A.; Filatov, V.L.; Kozlovsky, S.V.; Shilov, E.S.; Datskevich, P.N.; Rozov, F.N. Optimized dual assay for the transgenes selection and screening in CHO cell line development for recombinant protein production. Biotechnol. Lett. 2019, 41, 929–939. [Google Scholar] [CrossRef] [PubMed]
- Porter, A.J.; Racher, A.J.; Preziosi, R.; Dickson, A.J. Strategies for selecting recombinant CHO cell lines for cGMP manufacturing: Improving the efficiency of cell line generation. Biotechnol. Prog. 2010, 26, 1455–1464. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Albanetti, T.; Miro-Quesada, G.; Flack, L.; Li, L.; Klover, J.; Burson, K.; Evans, K.; Ivory, W.; Bowen, M.; et al. High-throughput screening of antibody-expressing CHO clones using an automated shaken deep-well system. Biotechnol. Prog. 2018, 34, 1460–1471. [Google Scholar] [CrossRef] [Green Version]
- Narayanan, H.; Luna, M.F.; von Stosch, M.; Bournazou, M.N.C.; Polotti, G.; Morbidelli, M.; Butté, A.; Sokolov, M. Bioprocessing in the Digital Age—The Role of Process Models. Biotechnol. J. 2019. [Google Scholar] [CrossRef]
- Xie, X.; Schenkendorf, R. Robust Process Design in Pharmaceutical Manufacturing under Batch-to-Batch Variation. Processes 2019, 7, 509. [Google Scholar] [CrossRef] [Green Version]
- Frahm, B. Seed Train Optimization for Cell Culture. In Animal Cell Biotechnology; Pörtner, R., Ed.; Springer protocols, Humana Press: New York, NY, USA, 2014; pp. 355–367. [Google Scholar]
- Hernández Rodríguez, T.; Posch, C.; Schmutzhard, J.; Stettner, J.; Weihs, C.; Pörtner, R.; Frahm, B. Predicting industrial-scale cell culture seed trains-A Bayesian framework for model fitting and parameter estimation, dealing with uncertainty in measurements and model parameters, applied to a nonlinear kinetic cell culture model, using an MCMC method. Biotechnol. Bioeng. 2019, 116, 2944–2959. [Google Scholar] [CrossRef] [Green Version]
- Hernández Rodríguez, T.; Frahm, B. Design, Optimization, and Adaptive Control of Cell Culture Seed Trains. Methods Mol. Biol. 2019, 2095, 251–267. [Google Scholar] [CrossRef]
- Hernández Rodríguez, T.; Frahm, B. Digital Seed Train Twins and Statistical Methods. Adv. Biochem. Eng. Biotechnol. 2020. [Google Scholar] [CrossRef]
- Deppe, S.; Frahm, B.; Hass, V.C.; Hernández Rodríguez, T.; Kuchemüller, K.B.; Möller, J.; Pörtner, R. Estimation of Process Model Parameters. Methods Mol. Biol. (Clifton, NJ) 2019, 2095, 213–234. [Google Scholar] [CrossRef]
- Zhang, X.; Stettler, M.; de Sanctis, D.; Perrone, M.; Parolini, N.; Discacciati, M.; de Jesus, M.; Hacker, D.; Quarteroni, A.; Wurm, F. Use of orbital shaken disposable bioreactors for mammalian cell cultures from the milliliter-scale to the 1,000-liter scale. Adv. Biochem. Eng. Biotechnol. 2009, 115, 33–53. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Bartlett, M.S. Properties of sufficiency and statistical tests. Proc. R. Soc. Lond. Ser. A Math. Phys. Sci. 1937, 160, 268–282. [Google Scholar] [CrossRef]
- Brown, M.B.; Forsythe, A.B. The Small Sample Behavior of Some Statistics Which Test the Equality of Several Means. Technometrics 1974, 16, 129–132. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kern, S.; Platas-Barradas, O.; Pörtner, R.; Frahm, B. Model-based strategy for cell culture seed train layout verified at lab scale. Cytotechnology 2016, 68, 1019–1032. [Google Scholar] [CrossRef] [Green Version]
- Möller, J.; Hernández Rodríguez, T.; Müller, J.; Arndt, L.; Kuchemüller, K.B.; Frahm, B.; Eibl, R.; Eibl, D.; Pörtner, R. Model uncertainty-based evaluation of process strategies during scale-up of biopharmaceutical processes. Comput. Chem. Eng. 2019, 106693. [Google Scholar] [CrossRef]
- MATLAB. Version 9.9.0 (R2020b); The MathWorks Inc.: Natick, MA, USA, 2010. [Google Scholar]
- Kim, Y.J.; Han, S.K.; Yoon, S.; Kim, C.W. Rich production media as a platform for CHO cell line development. AMB Express 2020, 10, 93. [Google Scholar] [CrossRef]
- Böhl, O.J.; Schellenberg, J.; Bahnemann, J.; Hitzmann, B.; Scheper, T.; Solle, D. Implementation of QbD strategies in the inoculum expansion of a mAb production process. Eng. Life Sci. 2020, 27, 9. [Google Scholar] [CrossRef]








| Scale | Cylindrical Vessel | Working Volume per Scale [L] |
|---|---|---|
| 1 | TubeSpin bioreactor 50 | 0.001–0.035 |
| 2 | Schott glass bottle (2 L) | 0.4–1.8 |
| 3 | Schott glass bottle (5 L) | 0.5–4.5 |
| 4 | OrbShake bioreactor prototype (50 L) | 15 |
| 5 | OrbShake bioreactor prototype (200 L) | 100 |
| 6 | OrbShake bioreactor prototype (2500 L) | 1000 |
| Balance Equations | Kinetic Equations |
|---|---|
| Biomass | |
| Substrates and metabolites | |
| Clone | Mean | sd | Max | cv | Mean | sd | Max | cv |
|---|---|---|---|---|---|---|---|---|
| [ h] | [%] | [ mg cell h] | [%] | |||||
| 1 | 0.028 | 0.0014 | 0.030 | 5 | 11.1 | 1.16 | 10.5 | 14.2 |
| 2 | 0.027 | 0.0019 | 0.030 | 7 | 17.4 | 1.01 | 19.6 | 5.8 |
| 3 | 0.026 | 0.0028 | 0.033 | 10 | 8.09 | 0.99 | 9.2 | 12.2 |
| 4 | 0.029 | 0.0018 | 0.033 | 6 | 9.92 | 0.76 | 10.8 | 7.6 |
| 5 | 0.030 | 0.0022 | 0.035 | 7 | 10.0 | 0.40 | 10.7 | 4.0 |
| Clones | p-Value | Clones | p-Value | ||
|---|---|---|---|---|---|
| [h ] | [-] | [1 mg cell h] | [-] | ||
| 5 vs. 3 | 0.0041 | 2 vs. 3 | 9.3 | ||
| 5 vs. 2 | 0.0039 | 2 vs. 4 | 7.5 | ||
| 4 vs. 3 | 0.0030 | 2 vs. 5 | 7.4 | ||
| 4 vs. 2 | 0.0028 | 2 vs. 1 | 6.4 | ||
| 5 vs. 1 | 0.0023 | 1 vs. 3 | 3.0 | ||
| 1 vs. 3 | 0.0018 | 5 vs. 3 | 1.9 | ||
| 1 vs. 2 | 0.0015 | 4 vs. 3 | 1.8 | ||
| 4 vs. 1 | 0.0012 | 1 vs. 4 | 1.1 | ||
| 5 vs. 4 | 0.0011 | 1 vs. 5 | 1.0 | ||
| 2 vs. 3 | 0.0003 | 1 | 5 vs. 4 | 0.1 | 1 |
| Parameter | Mean | Variance | cv |
|---|---|---|---|
| 0.0428 h | 5.36 h | 17% | |
| mg cell h | mg cell h | 33% |
| Clone | Remark | ||
|---|---|---|---|
| [h] | [ mg cellh] | ||
| A | reference clone | 0.042 | 3.90 |
| B | 7.6% lower , | 0.039 | 4.31 |
| 10.5% higher | |||
| C | 10% lower , | 0.038 | 6.90 |
| 74% higher |
| Response Variable | Factor | R | Coefficients () | Standard Error (SE) | p-Value |
|---|---|---|---|---|---|
| Volumetric titer (50 h) | 0.989 | ||||
| 0.42 | 0.016 | ||||
| 0.57 | 0.012 | ||||
| Overall process productivity (50 h) | 0.991 | ||||
| 0.61 | 0.013 | ||||
| 0.38 | 0.010 | ||||
| Volumetric titer (100 h) | 0.992 | ||||
| 0.69 | 0.013 | ||||
| 0.31 | 0.010 | ||||
| Overall process productivity (100 h) | 0.991 | ||||
| 0.75 | 0.014 | ||||
| 0.25 | 0.011 | ||||
| Volumetric titer (168 h) | 0.998 | ||||
| 0.33 | 0.008 | ||||
| 0.67 | 0.006 | ||||
| Overall process productivity (168 h) | 0.996 | ||||
| 0.54 | 0.009 | ||||
| 0.46 | 0.007 |
| Clone | Remark | [h] | [ mg cellh] | Volumetric Titer [mg L] | Overall Process Productivity [mg L h] |
|---|---|---|---|---|---|
| A | reference clone | 0.042 | 3.9 | 539 | 1.16 |
| B | 7.6% lower , | 0.039 | 4.31 | 558 | 1.14 |
| 10.5% higher | |||||
| C | 10% lower , | 0.0378 | 6.9 | 876 | 1.75 |
| 74% higher |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández Rodríguez, T.; Morerod, S.; Pörtner, R.; Wurm, F.M.; Frahm, B. Considerations of the Impacts of Cell-Specific Growth and Production Rate on Clone Selection—A Simulation Study. Processes 2021, 9, 964. https://doi.org/10.3390/pr9060964
Hernández Rodríguez T, Morerod S, Pörtner R, Wurm FM, Frahm B. Considerations of the Impacts of Cell-Specific Growth and Production Rate on Clone Selection—A Simulation Study. Processes. 2021; 9(6):964. https://doi.org/10.3390/pr9060964
Chicago/Turabian StyleHernández Rodríguez, Tanja, Sophie Morerod, Ralf Pörtner, Florian M. Wurm, and Björn Frahm. 2021. "Considerations of the Impacts of Cell-Specific Growth and Production Rate on Clone Selection—A Simulation Study" Processes 9, no. 6: 964. https://doi.org/10.3390/pr9060964
APA StyleHernández Rodríguez, T., Morerod, S., Pörtner, R., Wurm, F. M., & Frahm, B. (2021). Considerations of the Impacts of Cell-Specific Growth and Production Rate on Clone Selection—A Simulation Study. Processes, 9(6), 964. https://doi.org/10.3390/pr9060964