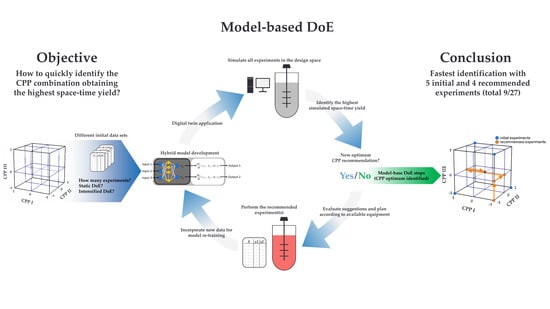
Digital Twin Application for Model-Based DoE to Rapidly Identify Ideal Process Conditions for Space-Time Yield Optimization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Data Sets
- Full factorial DoE: the fully characterized design space, used as a reference (N = 27)
- Fractional factorial DoE: the center point and the eight corners of the design space (N = 9)
- Fractional factorial DoE: the center point and four corners of the design space (N = 5)
- Fractional factorial DoE: the center point and two corners of the design space (N = 3)
- Complete iDoE: all iDoE cultivations, covering the entire design space (N = 9)
- Fractional iDoEs: one iDoE cultivation per induction level (N = 3, three different assemblies)
2.3. Hybrid Model Development
2.3.1. Model Building
2.3.2. Model Validation
2.3.3. Model Averaging
2.4. Digital Twin Application
3. Results
3.1. Analytical Space-Time Yield Maxima in the Design Space
3.2. Initial Training Data for the Model-Based DoE
3.3. Digital Twin Simulations of the Model-Based DoE
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ANN | artificial neural network |
| CMA | critical material attribute |
| CPP | critical process parameter |
| CQA | critical quality attribute |
| DoE | design of experiments |
| iDoE | intensified design of experiments |
| FDA | US federal drug administration |
| NRMSE | normalized root mean square error |
| PAT | process analytical technology |
| PI | prediction interval |
| QbD | quality by design |
| SD | standard deviation |
Appendix A
Appendix A.1. CPP Settings of All Experiments Used for Model-Based DoE
| CPP Combination | CPP 1 (µ) | CPP 2 (T) | CPP 3 (I) | Maximum Biomass (g L−1) | Maximum Space-Time Yield (g L−1 h−1) |
|---|---|---|---|---|---|
| 1 | 30 | 0.2 | 33.18 | 0.0193 | |
| 2 | 34 | 0.2 | 31.12 | 0.0726 | |
| 3 | 37 | 0.2 | 30.31 | 0.0311 | |
| 4 | 30 | 0.5 | 29.88 | 0.0733 | |
| 5 | 0.10 | 34 | 0.5 | 23.96 | 0.0837 |
| 6 | 37 | 0.5 | 20.6 | 0.0621 | |
| 7 | 30 | 0.9 | 26.07 | 0.0800 | |
| 8 | 34 | 0.9 | 20.69 | 0.0915 | |
| 9 | 37 | 0.9 | 18.23 | 0.0432 | |
| 10 | 30 | 0.2 | 34.28 | 0.0264 | |
| 11 | 34 | 0.2 | 32.09 | 0.0415 | |
| 12 | 37 | 0.2 | 29.7 | 0.0430 | |
| 13 | 30 | 0.5 | 31.74 | 0.0564 | |
| 14 | 0.15 | 34 | 0.5 | 28.66 | 0.0997 |
| 15 | 37 | 0.5 | 24.06 | 0.0663 | |
| 16 | 30 | 0.9 | 26.89 | 0.0564 | |
| 17 | 34 | 0.9 | 25.17 | 0.0815 | |
| 18 | 37 | 0.9 | 21.62 | 0.0485 | |
| 19 | 30 | 0.2 | 34.51 | 0.0157 | |
| 20 | 34 | 0.2 | 33.68 | 0.0227 | |
| 21 | 37 | 0.2 | 32.93 | 0.0274 | |
| 22 | 30 | 0.5 | 31.49 | 0.0418 | |
| 23 | 0.20 | 34 | 0.5 | 30.97 | 0.0783 |
| 24 | 37 | 0.5 | 28.85 | 0.0578 | |
| 25 | 30 | 0.9 | 29.14 | 0.0518 | |
| 26 | 34 | 0.9 | 29.25 | 0.0818 | |
| 27 | 37 | 0.9 | 23.98 | 0.0513 |
| iDoE CPP Combination | CPP 1 (µ) | CPP 2 (T) | CPP 3 (I) | CPP Shift 1 | CPP Shift 2 |
|---|---|---|---|---|---|
| 1 | 37 | 0.2 | 37 °C to 34 °C 0.10 h−1 to 0.20 h−1 | 0.20 h−1 to 0.10 h−1 | |
| 2 | 0.10 | 30 | 0.5 | 30 °C to 34 °C | 34 °C to 37 °C 0.10 h−1 to 0.20 h−1 |
| 3 | 34 | 0.9 | 34 °C to 37 °C | 0.10 h−1 to 0.15 h−1 | |
| 4 | 37 | 0.2 | 37 °C to 30 °C 0.15 h−1 to 0.10 h−1 | 30 °C to 34 °C 0.10 h−1 to 0.15 h−1 | |
| 5 | 0.15 | 30 | 0.5 | 0.15 h−1 to 0.20 h−1 | 30 °C to 34 °C |
| 6 | 34 | 0.5 | 34 °C to 37 °C | 0.15 h−1 to 0.10 h−1 | |
| 7 | 30 | 0.2 | 30 °C to 37 °C | 37 °C to 30 °C 0.20 h−1 to 0.15 h−1 | |
| 8 | 0.20 | 37 | 0.9 | 37 °C to 34 °C 0.20 h−1 to 0.15 h−1 | 34 °C to 30 °C 0.15 h−1 to 0.20 h−1 |
| 9 | 34 | 0.9 | 34 °C to 30 °C 0.20 h−1 to 0.15 h−1 | 0.15 h−1 to 0.10 h−1 |
Appendix A.2. Progression of the Recommended Experiments by Each Model-Based DoE Approach

References
- Pekarsky, A.; Konopek, V.; Spadiut, O. The impact of technical failures during cultivation of an inclusion body process. Bioprocess Biosyst. Eng. 2019, 42, 1611–1624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guideline, I.H.T. International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use Pharmaceutical development Q8(R2). ICH Harmon. Tripart. Guidel. 2009, 1–24. [Google Scholar] [CrossRef]
- Mandenius, C.-F.; Graumann, K.; Schultz, T.W.; Premstaller, A.; Olsson, I.M.; Petiot, E.; Clemens, C.; Welin, M. Quality-by-design for biotechnology-related pharmaceuticals. Biotechnol. J. 2009, 4, 600–609. [Google Scholar] [CrossRef]
- Rathore, A.S.; Winkle, H. Quality by design for biopharmaceuticals. Nat. Biotechnol. 2009, 27, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Lundstedt, T.; Seifert, E.; Abramo, L.; Thelin, B.; Nyström, Å.; Pettersen, J.; Bergman, R. Experimental design and optimization. Chemom. Intell. Lab. Syst. 1998, 42, 3–40. [Google Scholar] [CrossRef]
- Mandenius, C.-F.; Brundin, A. Bioprocess Optimization, Using Design-of-experiments Methodology. Biotechnol. Progr. 2008, 24, 1191–1203. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-M.; Gilmore, D.F. Statistical Experimental Design for Bioprocess Modeling and Optimization Analysis. Appl. Biochem. Biotechnol. 2006, 135, 101–135. [Google Scholar] [CrossRef]
- Hallow, D.M.; Mudryk, B.M.; Braem, A.D.; Tabora, J.E.; Lyngberg, O.K.; Bergum, J.S.; Rossano, L.T.; Tummala, S. An example of utilizing mechanistic and empirical modeling in quality by design. J. Pharm. Innov. 2010, 5, 193–203. [Google Scholar] [CrossRef]
- Kadlec, P.; Gabrys, B.; Strandt, S. Data-driven Soft Sensors in the process industry. Comput. Chem. Eng. 2009, 33, 795–814. [Google Scholar] [CrossRef] [Green Version]
- von Stosch, M.; Davy, S.; Francois, K.; Galvanauskas, V.; Hamelink, J.M.; Luebbert, A.; Mayer, M.; Oliveira, R.; O’Kennedy, R.; Rice, P.; et al. Hybrid modeling for quality by design and PAT-benefits and challenges of applications in biopharmaceutical industry. Biotechnol. J. 2014, 9, 719–726. [Google Scholar] [CrossRef] [Green Version]
- Bayer, B.; Von Stosch, M.; Striedner, G.; Duerkop, M. Comparison of Modeling Methods for DoE-Based Holistic Upstream Process Characterization. Biotechnol. J. 2020, 15. [Google Scholar] [CrossRef] [Green Version]
- Taylor, P.; Picard, R.R.; Cook, R.D. Cross-Validation of Regression Models. J. Am. Stat. Assoc. 1984, 79, 575–583. [Google Scholar] [CrossRef]
- Mendes-Moreira, J.; Soares, C.; Jorge, A.M.; De Sousa, J.F. Ensemble approaches for regression: A survey. ACM Comput. Surv. 2012, 45. [Google Scholar] [CrossRef]
- von Stosch, M.; Oliveira, R.; Peres, J.; Feyo de Azevedo, S. Hybrid semi-parametric modeling in process systems engineering: Past, present and future. Comput. Chem. Eng. 2014, 60, 86–101. [Google Scholar] [CrossRef] [Green Version]
- Krippl, M.; Dürauer, A.; Duerkop, M. Hybrid modeling of cross-flow filtration: Predicting the flux evolution and duration of ultrafiltration processes. Sep. Purif. Technol. 2020, 248, 1–11. [Google Scholar] [CrossRef]
- Krippl, M.; Bofarull-Manzano, I.; Duerkop, M.; Dürauer, A. Hybrid modeling for simultaneous prediction of flux, rejection factor and concentration in two-component crossflow ultrafiltration. Processes 2020, 8, 1625. [Google Scholar] [CrossRef]
- Wang, G.; Briskot, T.; Hahn, T.; Baumann, P.; Hubbuch, J. Estimation of adsorption isotherm and mass transfer parameters in protein chromatography using artificial neural networks. J. Chromatogr. A 2017, 1487, 211–217. [Google Scholar] [CrossRef]
- Kalil, S.J.; Maugeri, F.; Rodrigues, M.I. Response surface analysis and simulation as a tool for bioprocess design and optimization. Process Biochem. 2000, 35, 539–550. [Google Scholar] [CrossRef]
- Sommeregger, W.; Sissolak, B.; Kandra, K.; von Stosch, M.; Mayer, M.; Striedner, G. Quality by control: Towards model predictive control of mammalian cell culture bioprocesses. Biotechnol. J. 2017, 12. [Google Scholar] [CrossRef] [Green Version]
- Schmidberger, T.; Gutmann, R.; Bayer, K.; Kronthaler, J.; Huber, R. Advanced online monitoring of cell culture off-gas using proton transfer reaction mass spectrometry. Biotechnol. Prog. 2013, 7. [Google Scholar] [CrossRef]
- Bayer, B.; Von Stosch, M.; Melcher, M.; Duerkop, M.; Striedner, G. Soft sensor based on 2D-fluorescence and process data enabling real-time estimation of biomass in Escherichia coli cultivations. Eng. Life Sci. 2020, 20, 26–35. [Google Scholar] [CrossRef] [Green Version]
- Luttmann, R.; Bracewell, D.G.; Cornelissen, G.; Gernaey, K.V.; Glassey, J.; Hass, V.C.; Kaiser, C.; Preusse, C.; Striedner, G.; Mandenius, C.-F. Soft sensors in bioprocessing: A status report and recommendations. Biotechnol. J. 2012, 7, 1040–1048. [Google Scholar] [CrossRef]
- Morari, M.; Lee, J.H. Model predictive control: Past, present and future. Comput. Chem. Eng. 1999, 23, 667–682. [Google Scholar] [CrossRef]
- Kroll, P.; Hofer, A.; Ulonska, S.; Kager, J.; Herwig, C. Model-Based Methods in the Biopharmaceutical Process Lifecycle. Pharm. Res. 2017, 34, 2596–2613. [Google Scholar] [CrossRef] [Green Version]
- Udugama, I.A.; Lopez, P.C.; Gargalo, C.L.; Li, X.; Bayer, C.; Gernaey, K.V. Digital Twin in biomanufacturing: Challenges and opportunities towards its implementation. Syst. Microbiol. Biomanuf. 2021. [Google Scholar] [CrossRef]
- Kritzinger, W.; Karner, M.; Traar, G.; Henjes, J.; Sihn, W. Digital Twin in manufacturing: A categorical literature review and classification. IFAC-PapersOnLine 2018, 51, 1016–1022. [Google Scholar] [CrossRef]
- Shahmohammadi, A.; McAuley, K.B. Using prior parameter knowledge in model-based design of experiments for pharmaceutical production. AIChE J. 2020, 66. [Google Scholar] [CrossRef]
- Abt, V.; Barz, T.; Cruz, N.; Herwig, C.; Kroll, P.; Möller, J.; Pörtner, R.; Schenkendorf, R. Model-based tools for optimal experiments in bioprocess engineering. Curr. Opin. Chem. Eng. 2018, 22, 244–252. [Google Scholar] [CrossRef]
- Smiatek, J.; Jung, A.; Bluhmki, E. Towards a Digital Bioprocess Replica: Computational Approaches in Biopharmaceutical Development and Manufacturing. Trends Biotechnol. 2020, 38, 1141–1153. [Google Scholar] [CrossRef]
- Möller, J.; Kuchemüller, K.B.; Steinmetz, T.; Koopmann, K.S.; Pörtner, R. Model-assisted Design of Experiments as a concept for knowledge-based bioprocess development. Bioprocess Biosyst. Eng. 2019, 42, 867–882. [Google Scholar] [CrossRef]
- Narayanan, H.; Luna, M.F.; von Stosch, M.; Cruz Bournazou, M.N.; Polotti, G.; Morbidelli, M.; Butté, A.; Sokolov, M. Bioprocessing in the Digital Age: The Role of Process Models. Biotechnol. J. 2020, 15, 1–10. [Google Scholar] [CrossRef]
- von Stosch, M.; Willis, M.J. Intensified Design of Experiments for upstream bioreactors. Eng. Life Sci. 2016, 17, 1173–1184. [Google Scholar] [CrossRef]
- Cserjan-Puschmann, M.; Kramer, W.; Duerrschmid, E.; Striedner, G.; Bayer, K. Metabolic approaches for the optimisation of recombinant fermentation processes. Appl. Microbiol. Biotechnol. 1999, 53, 43–50. [Google Scholar] [CrossRef]
- Porstmann, T.; Wietschke, R.; Schmechta, H.; Grunow, R.; Porstmann, B.; Bleiber, R.; Pergande, M.; Stachat, S.; von Baehr, R. A rapid and sensitive enzyme immunoassay for Cu/Zn superoxide dismutase with polyclonal and monoclonal antibodies. Clin. Chim. Acta 1988, 171, 1–10. [Google Scholar] [CrossRef]
- Marisch, K.; Bayer, K.; Cserjan-Puschmann, M.; Luchner, M.; Striedner, G. Evaluation of three industrial Escherichia coli strains in fed-batch cultivations during high-level SOD protein production. Microb. Cell Fact. 2013, 12, 58. [Google Scholar] [CrossRef] [Green Version]
- Luchner, M.; Striedner, G.; Cserjan-Puschmann, M.; Strobl, F.; Bayer, K. Online prediction of product titer and solubility of recombinant proteins in Escherichia coli fed-batch cultivations. J. Chem. Technol. Biotechnol. 2015, 90, 283–290. [Google Scholar] [CrossRef]
- Melcher, M.; Scharl, T.; Spangl, B.; Luchner, M.; Cserjan, M.; Bayer, K.; Leisch, F.; Striedner, G. The potential of random forest and neural networks for biomass and recombinant protein modeling in Escherichia coli fed-batch fermentations. Biotechnol. J. 2015, 10, 1770–1782. [Google Scholar] [CrossRef] [PubMed]
- Bayer, B.; Striedner, G.; Duerkop, M. Hybrid Modeling and Intensified DoE: An Approach to Accelerate Upstream Process Characterization. Biotechnol. J. 2020, 15. [Google Scholar] [CrossRef] [PubMed]
- Mercier, S.M.; Diepenbroek, B.; Wijffels, R.H.; Streefland, M. Multivariate PAT solutions for biopharmaceutical cultivation: Current progress and limitations. Trends Biotechnol. 2014, 32, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Cardillo, A.G.; Castellanos, M.M.; Desailly, B.; Dessoy, S.; Mariti, M.; Portela, R.M.C.; Scutella, B.; von Stosch, M.; Tomba, E.; Varsakelis, C. Towards in silico Process Modeling for Vaccines. Trends Biotechnol. 2021, 1–11. [Google Scholar] [CrossRef]




| Digital Twin Conversion | CPP I (µ) | CPP II (T) | CPP III (I) | Analytical Maximum (g L−1 h−1) | Simulated Maximum (g L−1 h−1) |
|---|---|---|---|---|---|
| 1st recommendation | 0.10 | 30 | 0.2 | 0.0185 (±0.0006) | 0.1605 (±0.0185) |
| 2nd recommendation | 0.10 | 30 | 0.5 | 0.0696 (±0.0029) | 0.1220 (±0.0058) |
| 3rd recommendation | 0.10 | 34 | 0.5 | 0.0820 (±0.0018) | 0.1303 (±0.0040) |
| 4th recommendation | 0.20 | 34 | 0.5 | 0.0755 (±0.0032) | 0.0848 (±0.0079) |
| 5th recommendation | 0.15 | 34 | 0.5 | 0.0976 (±0.0026) | 0.0955 (±0.0186) |
| Initial Data Set | Initial Experiments | Recommended Experiments | Total Experiments | Optimum Found |
|---|---|---|---|---|
| full factorial DoE (A) | 27 | 0 | 27 | yes |
| fractional factorial DoE (B) | 9 | 2 | 11 | yes |
| fractional factorial DoE (C) | 5 | 4 | 9 | yes |
| fractional factorial DoE (D) | 3 | 7 | 10 | yes |
| complete iDoE (E) | 9 | 2 | 11 | no |
| fractional iDoEs (F–H) | 3 | 1–4 | 4–7 | no |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bayer, B.; Dalmau Diaz, R.; Melcher, M.; Striedner, G.; Duerkop, M. Digital Twin Application for Model-Based DoE to Rapidly Identify Ideal Process Conditions for Space-Time Yield Optimization. Processes 2021, 9, 1109. https://doi.org/10.3390/pr9071109
Bayer B, Dalmau Diaz R, Melcher M, Striedner G, Duerkop M. Digital Twin Application for Model-Based DoE to Rapidly Identify Ideal Process Conditions for Space-Time Yield Optimization. Processes. 2021; 9(7):1109. https://doi.org/10.3390/pr9071109
Chicago/Turabian StyleBayer, Benjamin, Roger Dalmau Diaz, Michael Melcher, Gerald Striedner, and Mark Duerkop. 2021. "Digital Twin Application for Model-Based DoE to Rapidly Identify Ideal Process Conditions for Space-Time Yield Optimization" Processes 9, no. 7: 1109. https://doi.org/10.3390/pr9071109
APA StyleBayer, B., Dalmau Diaz, R., Melcher, M., Striedner, G., & Duerkop, M. (2021). Digital Twin Application for Model-Based DoE to Rapidly Identify Ideal Process Conditions for Space-Time Yield Optimization. Processes, 9(7), 1109. https://doi.org/10.3390/pr9071109