Graphene-Based Electrochemical Nano-Biosensors for Detection of SARS-CoV-2
Abstract
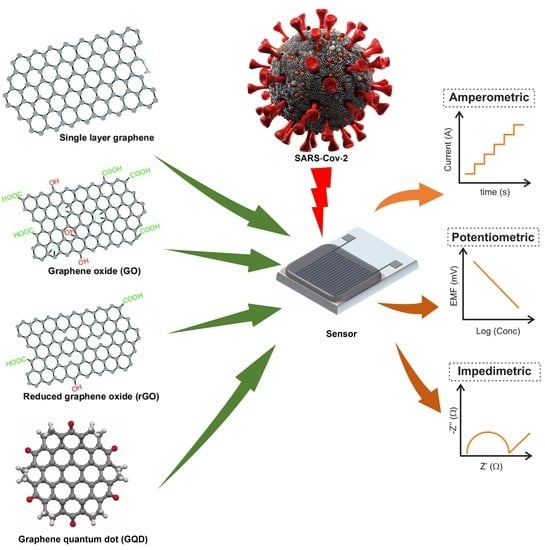
:1. Introduction
1.1. SARS-CoV-2 and Biosensors
- Transmission: SARS-CoV-2 is primarily transmitted through respiratory droplets when an infected person coughs, sneezes, or talks [1]. The virus can also be spread by touching a surface contaminated with the virus and then touching one’s face. Airborne transmission is also possible in certain settings, particularly enclosed spaces with poor ventilation [2].
- Incubation period: The incubation period for SARS-CoV-2 ranges from 2 to 14 days, with an average of 5 to 6 days. However, some people may develop symptoms outside of this range [3].
- Symptoms: The most common symptoms of COVID-19 include fever, cough, and shortness of breath. Other symptoms may include fatigue, body aches, headache, loss of smell or taste, sore throat, congestion, and runny nose. Some people may be asymptomatic, meaning they do not have any symptoms.
- Severity: COVID-19 can range in severity from mild to severe illness and can be fatal in some cases. Older adults and people with underlying health conditions, such as diabetes, obesity, heart disease, or weakened immune systems, are at a higher risk for severe illness and death [4].
- Case fatality rate: The case fatality rate (CFR) for COVID-19 varies by age and underlying health conditions. The overall global CFR has been estimated to be around 0.9% as of February 2023 [5].
- Variants: SARS-CoV-2 has mutated over time, leading to the emergence of several variants of concern (VOCs) and variants of interest (VOIs). VOCs include the Alpha, Beta, Gamma, and Delta variants, which are believed to be more transmissible and potentially more severe than the original strain of the virus. VOIs include several other variants with mutations that may impact transmission, severity, or immune response [6].
- Global impact: Since the start of the pandemic, COVID-19 has spread to virtually every country in the world, causing significant morbidity and mortality (Figure 1). As of 17 March 2023, there have been over 760 million confirmed cases and over 6.8 million deaths reported globally [5]. The impact of the pandemic has also had significant economic and social consequences, including disruptions to healthcare systems, education, and employment [7].
- Rapid detection: Biosensors can provide results in minutes or even seconds, which is crucial for timely diagnosis and treatment.
- High sensitivity: Biosensors can detect very low concentrations of the virus, which is important for early detection and surveillance.
- Low cost: Biosensors can be produced at a lower cost than traditional methods, making them more accessible in resource-limited settings.
- Portable: Biosensors can be designed to be portable, allowing for point-of-care testing in remote or underserved areas.
- Non-invasive: Biosensors can detect the virus from various sources, including saliva, urine, and breath, without the need for invasive sampling methods.
1.2. Graphene in Biosensor
- High surface area and conductivity allow efficient capture and transduction of biomolecular interactions [26].
- High optical transparency and tunability enable optical sensing methods such as fluorescence, Raman scattering, and plasmon resonance [27].
- High mechanical flexibility and stability enable the fabrication of various device architectures, such as crumpled graphene or graphene nanoribbons [28].
2. Electrochemical Biosensors for the Detection of SARS-CoV-2
2.1. Amperometric Biosensors for the Detection of SARS-CoV-2
2.1.1. Using Graphene
2.1.2. Using Graphene Oxide
2.1.3. Using Reduced Graphene Oxide
2.1.4. Using Graphene Quantum Dots
2.2. Potentiometric Biosensors for the Detection of SARS-CoV-2
2.2.1. Using Graphene
2.2.2. Using Graphene Oxide
2.2.3. Using Reduced Graphene Oxide
2.3. Impedimetric Biosensors for the Detection of SARS-CoV-2
2.3.1. Using Graphene
2.3.2. Using Graphene Oxide
2.3.3. Using Reduced Graphene Oxide
3. Current Challenges and Future Perspectives
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Jayaweera, M.; Perera, H.; Gunawardana, B.; Manatunge, J. Transmission of COVID-19 Virus by Droplets and Aerosols: A Critical Review on the Unresolved Dichotomy. Environ. Res. 2020, 188, 109819. [Google Scholar] [CrossRef]
- Karia, R.; Gupta, I.; Khandait, H.; Yadav, A.; Yadav, A. COVID-19 and Its Modes of Transmission. SN Compr. Clin. Med. 2020, 2, 1798–1801. [Google Scholar] [CrossRef] [PubMed]
- Lauer, S.A.; Grantz, K.H.; Bi, Q.; Jones, F.K.; Zheng, Q.; Meredith, H.R.; Azman, A.S.; Reich, N.G.; Lessler, J. The Incubation Period of Coronavirus Disease 2019 (COVID-19) from Publicly Reported Confirmed Cases: Estimation and Application. Ann. Intern. Med. 2020, 172, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhong, X.; Wang, Y.; Zeng, X.; Luo, T.; Liu, Q. Clinical Determinants of the Severity of COVID-19: A Systematic Review and Meta-Analysis. PLoS ONE 2021, 16, e0250602. [Google Scholar] [CrossRef] [PubMed]
- WHO. Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int (accessed on 17 March 2023).
- Carabelli, A.M.; Peacock, T.P.; Thorne, L.G.; Harvey, W.T.; Hughes, J.; de Silva, T.I.; Peacock, S.J.; Barclay, W.S.; de Silva, T.I.; Towers, G.J.; et al. SARS-CoV-2 Variant Biology: Immune Escape, Transmission and Fitness. Nat. Rev. Microbiol. 2023, 21, 162–177. [Google Scholar] [CrossRef]
- Wei, X.; Li, L.; Zhang, F. The Impact of the COVID-19 Pandemic on Socio-Economic and Sustainability. Environ. Sci. Pollut. Res. 2021, 28, 68251–68260. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. COVID Data Tracker; US Department of Health and Human Services, CDC: Atlanta, GA, USA, 2022. Available online: https://covid.cdc.gov/covid-data-tracker (accessed on 11 April 2023).
- Wang, C.; Liu, Z.; Chen, Z.; Huang, X.; Xu, M.; He, T.; Zhang, Z. The Establishment of Reference Sequence for SARS-CoV-2 and Variation Analysis. J. Med. Virol. 2020, 92, 667–674. [Google Scholar] [CrossRef]
- Pizzato, M.; Baraldi, C.; Boscato Sopetto, G.; Finozzi, D.; Gentile, C.; Gentile, M.D.; Marconi, R.; Paladino, D.; Raoss, A.; Riedmiller, I.; et al. SARS-CoV-2 and the Host Cell: A Tale of Interactions. Front. Virol. 2022, 1, 815388. [Google Scholar] [CrossRef]
- Santos, I.d.A.; Grosche, V.R.; Bergamini, F.R.G.; Sabino-Silva, R.; Jardim, A.C.G. Antivirals Against Coronaviruses: Candidate Drugs for SARS-CoV-2 Treatment? Front. Microbiol. 2020, 11, 1818. [Google Scholar] [CrossRef]
- Rahman, M.M. Progress in Electrochemical Biosensing of SARS-CoV-2 Virus for COVID-19 Management. Chemosensors 2022, 10, 287. [Google Scholar] [CrossRef]
- Cassedy, A.; Parle-McDermott, A.; O’Kennedy, R. Virus Detection: A Review of the Current and Emerging Molecular and Immunological Methods. Front. Mol. Biosci. 2021, 8, 637559. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, J.; Hussain, C.M. The Emergence of Carbon Nanomaterials as Effective Nano-Avenues to Fight against COVID-19. Materials 2023, 16, 1068. [Google Scholar] [CrossRef]
- Murillo, A.M.M.; Tomé-Amat, J.; Ramírez, Y.; Garrido-Arandia, M.; Valle, L.G.; Hernández-Ramírez, G.; Tramarin, L.; Herreros, P.; Santamaría, B.; Díaz-Perales, A.; et al. Developing an Optical Interferometric Detection Method Based Biosensor for Detecting Specific SARS-CoV-2 Immunoglobulins in Serum and Saliva, and Their Corresponding ELISA Correlation. Sens. Actuators B Chem. 2021, 345, 130394. [Google Scholar] [CrossRef]
- Kaushik, A.K.; Dhau, J.S.; Gohel, H.; Mishra, Y.K.; Kateb, B.; Kim, N.-Y.; Goswami, D.Y. Electrochemical SARS-CoV-2 Sensing at Point-of-Care and Artificial Intelligence for Intelligent COVID-19 Management. ACS Appl. Bio Mater. 2020, 3, 7306–7325. [Google Scholar] [CrossRef] [PubMed]
- Mandal, D.; Indaleeb, M.M.; Younan, A.; Banerjee, S. Piezoelectric Point-of-Care Biosensor for the Detection of SARS-COV-2 (COVID-19) Antibodies. Sens. Bio Sens. Res. 2022, 37, 100510. [Google Scholar] [CrossRef]
- Zhang, J.Z.; Yeh, H.-W.; Walls, A.C.; Wicky, B.I.M.; Sprouse, K.R.; VanBlargan, L.A.; Treger, R.; Quijano-Rubio, A.; Pham, M.N.; Kraft, J.C.; et al. Thermodynamically Coupled Biosensors for Detecting Neutralizing Antibodies against SARS-CoV-2 Variants. Nat. Biotechnol. 2022, 40, 1336–1340. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Saha, R.; Su, D.; Krishna, V.D.; Liu, J.; Cheeran, M.C.-J.; Wang, J.-P. Magnetic-Nanosensor-Based Virus and Pathogen Detection Strategies before and during COVID-19. ACS Appl. Nano Mater. 2020, 3, 9560–9580. [Google Scholar] [CrossRef]
- Guliy, O.; Zaitsev, B.; Teplykh, A.; Balashov, S.; Fomin, A.; Staroverov, S.; Borodina, I. Acoustical Slot Mode Sensor for the Rapid Coronaviruses Detection. Sensors 2021, 21, 1822. [Google Scholar] [CrossRef] [PubMed]
- Drobysh, M.; Ramanaviciene, A.; Viter, R.; Chen, C.-F.; Samukaite-Bubniene, U.; Ratautaite, V.; Ramanavicius, A. Biosensors for the Determination of SARS-CoV-2 Virus and Diagnosis of COVID-19 Infection. Int. J. Mol. Sci. 2022, 23, 666. [Google Scholar] [CrossRef]
- Chaibun, T.; Puenpa, J.; Ngamdee, T.; Boonapatcharoen, N.; Athamanolap, P.; O’Mullane, A.P.; Vongpunsawad, S.; Poovorawan, Y.; Lee, S.Y.; Lertanantawong, B. Rapid Electrochemical Detection of Coronavirus SARS-CoV-2. Nat. Commun. 2021, 12, 802. [Google Scholar] [CrossRef]
- Rasmi, Y.; Li, X.; Khan, J.; Ozer, T.; Choi, J.R. Emerging Point-of-Care Biosensors for Rapid Diagnosis of COVID-19: Current Progress, Challenges, and Future Prospects. Anal. Bioanal. Chem. 2021, 413, 4137–4159. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, J.; Hussain, C.M. Decadal Journey of CNT-Based Analytical Biosensing Platforms in the Detection of Human Viruses. Nanomaterials 2022, 12, 4132. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, J.; Hussain, C.M. Graphene and Its Derivatives for Analytical Lab on Chip Platforms. TrAC Trends Anal. Chem. 2019, 114, 326–337. [Google Scholar] [CrossRef]
- Kamedulski, P.; Skorupska, M.; Binkowski, P.; Arendarska, W.; Ilnicka, A.; Lukaszewicz, J.P. High Surface Area Micro-Mesoporous Graphene for Electrochemical Applications. Sci. Rep. 2021, 11, 22054. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Gan, X.; Jia, B.; Mao, D.; Zhao, J. Tunable High-Efficiency Light Absorption of Monolayer Graphene via Tamm Plasmon Polaritons. Opt. Lett. OL 2016, 41, 4743–4746. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Choi, K.; Lee, B.; Kim, Y.; Hong, B.H. Materials for Flexible, Stretchable Electronics: Graphene and 2D Materials. Annu. Rev. Mater. Res. 2015, 45, 63–84. [Google Scholar] [CrossRef]
- Catania, F.; Marras, E.; Giorcelli, M.; Jagdale, P.; Lavagna, L.; Tagliaferro, A.; Bartoli, M. A Review on Recent Advancements of Graphene and Graphene-Related Materials in Biological Applications. Appl. Sci. 2021, 11, 614. [Google Scholar] [CrossRef]
- Reghunath, R.; Devi, K.; Singh, K.K. Recent Advances in Graphene Based Electrochemical Glucose Sensor. Nano-Struct. Nano-Objects 2021, 26, 100750. [Google Scholar] [CrossRef]
- Arumugasamy, S.K.; Govindaraju, S.; Yun, K. Electrochemical Sensor for Detecting Dopamine Using Graphene Quantum Dots Incorporated with Multiwall Carbon Nanotubes. Appl. Surf. Sci. 2020, 508, 145294. [Google Scholar] [CrossRef]
- Wu, S.; Jiang, M.; Mao, H.; Zhao, N.; He, D.; Chen, Q.; Liu, D.; Zhang, W.; Song, X.-M. A Sensitive Cholesterol Electrochemical Biosensor Based on Biomimetic Cerasome and Graphene Quantum Dots. Anal. Bioanal. Chem. 2022, 414, 3593–3603. [Google Scholar] [CrossRef]
- Wang, C.-F.; Sun, X.-Y.; Su, M.; Wang, Y.-P.; Lv, Y.-K. Electrochemical Biosensors Based on Antibody, Nucleic Acid and Enzyme Functionalized Graphene for the Detection of Disease-Related Biomolecules. Analyst 2020, 145, 1550–1562. [Google Scholar] [CrossRef] [PubMed]
- Fritea, L.; Tertis, M.; Sandulescu, R.; Cristea, C. Chapter Eleven—Enzyme–Graphene Platforms for Electrochemical Biosensor Design With Biomedical Applications. In Methods in Enzymology; Kumar, C.V., Ed.; Enzyme Nanoarchitectures: Enzymes Armored with Graphene; Academic Press: Cambridge, MA, USA, 2018; Volume 609, pp. 293–333. [Google Scholar]
- Bai, Y.; Xu, T.; Zhang, X. Graphene-Based Biosensors for Detection of Biomarkers. Micromachines 2020, 11, 60. [Google Scholar] [CrossRef] [PubMed]
- Georgakilas, V.; Otyepka, M.; Bourlinos, A.B.; Chandra, V.; Kim, N.; Kemp, K.C.; Hobza, P.; Zboril, R.; Kim, K.S. Functionalization of Graphene: Covalent and Non-Covalent Approaches, Derivatives and Applications. Chem. Rev. 2012, 112, 6156–6214. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Raya, J.; Ji, D.; Nishina, Y.; Ménard-Moyon, C.; Bianco, A. Is Carboxylation an Efficient Method for Graphene Oxide Functionalization? Nanoscale Adv. 2020, 2, 4085–4092. [Google Scholar] [CrossRef] [PubMed]
- Sierra, U.; Cuara, E.; Mercado, A.; Díaz-Barriga, E.; Bahena, A.; Cortés, A.; Martínez, J.P.; Solà, M.; Fernández, S. Efficient Synthesis of Amine-Functionalized Graphene Oxide by Ultrasound-Assisted Reactions and Density Functional Theory Mechanistic Insight. Appl. Nanosci. 2021, 11, 1637–1649. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, P.; Fan, J.; Yu, H. Covalently Functionalized Graphene by Thiourea for Enhancing H2-Evolution Performance of TiO2 Photocatalyst. Ceram. Int. 2021, 47, 654–661. [Google Scholar] [CrossRef]
- Yu, W.; Sisi, L.; Haiyan, Y.; Jie, L. Progress in the Functional Modification of Graphene/Graphene Oxide: A Review. RSC Adv. 2020, 10, 15328–15345. [Google Scholar] [CrossRef]
- Georgakilas, V.; Tiwari, J.N.; Kemp, K.C.; Perman, J.A.; Bourlinos, A.B.; Kim, K.S.; Zboril, R. Noncovalent Functionalization of Graphene and Graphene Oxide for Energy Materials, Biosensing, Catalytic, and Biomedical Applications. Chem. Rev. 2016, 116, 5464–5519. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.-Y.; Chen, W.-H.; Huang, C.-H. Chapter 15—Graphene in Electrochemical Biosensors. In Biomedical Applications of Graphene and 2D Nanomaterials; Nurunnabi, M., McCarthy, J.R., Eds.; Micro and Nano Technologies; Elsevier: Amsterdam, The Netherlands, 2019; pp. 321–336. ISBN 978-0-12-815889-0. [Google Scholar]
- Perreault, F.; Faria, A.F.d.; Elimelech, M. Environmental Applications of Graphene-Based Nanomaterials. Chem. Soc. Rev. 2015, 44, 5861–5896. [Google Scholar] [CrossRef]
- Gosai, A.; Khondakar, K.R.; Ma, X.; Ali, M.A. Application of Functionalized Graphene Oxide Based Biosensors for Health Monitoring: Simple Graphene Derivatives to 3D Printed Platforms. Biosensors 2021, 11, 384. [Google Scholar] [CrossRef]
- Peña-Bahamonde, J.; Nguyen, H.N.; Fanourakis, S.K.; Rodrigues, D.F. Recent Advances in Graphene-Based Biosensor Technology with Applications in Life Sciences. J. Nanobiotechnol. 2018, 16, 75. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Park, S.-J.; Choi, J.-W. Electrical Property of Graphene and Its Application to Electrochemical Biosensing. Nanomaterials 2019, 9, 297. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, J.; Hussain, C.M. Graphene-Based Field-Effect Transistor Biosensors for the Rapid Detection and Analysis of Viruses: A Perspective in View of COVID-19. Carbon. Trends 2021, 2, 100011. [Google Scholar] [CrossRef]
- Alhazmi, H.A.; Ahsan, W.; Mangla, B.; Javed, S.; Hassan, M.Z.; Asmari, M.; Bratty, M.A.; Najmi, A. Graphene-Based Biosensors for Disease Theranostics: Development, Applications, and Recent Advancements. Nanotechnol. Rev. 2022, 11, 96–116. [Google Scholar] [CrossRef]
- Liang, X.; Li, N.; Zhang, R.; Yin, P.; Zhang, C.; Yang, N.; Liang, K.; Kong, B. Carbon-Based SERS Biosensor: From Substrate Design to Sensing and Bioapplication. NPG Asia Mater. 2021, 13, 1–36. [Google Scholar] [CrossRef]
- Sengupta, J.; Adhikari, A.; Hussain, C.M. Graphene-Based Analytical Lab-on-Chip Devices for Detection of Viruses: A Review. Carbon. Trends 2021, 4, 100072. [Google Scholar] [CrossRef]
- Ansari, A.A.; Malhotra, B.D. Current Progress in Organic–Inorganic Hetero-Nano-Interfaces Based Electrochemical Biosensors for Healthcare Monitoring. Coord. Chem. Rev. 2022, 452, 214282. [Google Scholar] [CrossRef]
- Erdem, A.; Senturk, H.; Yildiz, E.; Maral, M. Amperometric Immunosensor Developed for Sensitive Detection of SARS-CoV-2 Spike S1 Protein in Combined with Portable Device. Talanta 2022, 244, 123422. [Google Scholar] [CrossRef] [PubMed]
- Ghanam, A.; Mohammadi, H.; Amine, A.; Haddour, N.; Buret, F. Chemical Sensors. In Encyclopedia of Sensors and Biosensors, 1st ed.; Narayan, R., Ed.; Elsevier: Oxford, UK, 2023; pp. 161–177. ISBN 978-0-12-822549-3. [Google Scholar]
- Torrente-Rodríguez, R.M.; Lukas, H.; Tu, J.; Min, J.; Yang, Y.; Xu, C.; Rossiter, H.B.; Gao, W. SARS-CoV-2 RapidPlex: A Graphene-Based Multiplexed Telemedicine Platform for Rapid and Low-Cost COVID-19 Diagnosis and Monitoring. Matter 2020, 3, 1981–1998. [Google Scholar] [CrossRef]
- Jaewjaroenwattana, J.; Phoolcharoen, W.; Pasomsub, E.; Teengam, P.; Chailapakul, O. Electrochemical Paper-Based Antigen Sensing Platform Using Plant-Derived Monoclonal Antibody for Detecting SARS-CoV-2. Talanta 2023, 251, 123783. [Google Scholar] [CrossRef]
- Ali, M.A.; Hu, C.; Zhang, F.; Jahan, S.; Yuan, B.; Saleh, M.S.; Gao, S.-J.; Panat, R. N Protein-Based Ultrasensitive SARS-CoV-2 Antibody Detection in Seconds via 3D Nanoprinted, Microarchitected Array Electrodes. J. Med. Virol. 2022, 94, 2067–2078. [Google Scholar] [CrossRef] [PubMed]
- Beduk, T.; Beduk, D.; de Oliveira Filho, J.I.; Zihnioglu, F.; Cicek, C.; Sertoz, R.; Arda, B.; Goksel, T.; Turhan, K.; Salama, K.N.; et al. Rapid Point-of-Care COVID-19 Diagnosis with a Gold-Nanoarchitecture-Assisted Laser-Scribed Graphene Biosensor. Anal. Chem. 2021, 93, 8585–8594. [Google Scholar] [CrossRef] [PubMed]
- Mojsoska, B.; Larsen, S.; Olsen, D.A.; Madsen, J.S.; Brandslund, I.; Alatraktchi, F.A. Rapid SARS-CoV-2 Detection Using Electrochemical Immunosensor. Sensors 2021, 21, 390. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Zeng, X.; Zhang, J.; Kong, J.; Fang, X. Accurate Identification of SARS-CoV-2 Variant Delta Using Graphene/CRISPR-DCas9 Electrochemical Biosensor. Talanta 2022, 249, 123687. [Google Scholar] [CrossRef] [PubMed]
- Damiati, S.; Søpstad, S.; Peacock, M.; Akhtar, A.S.; Pinto, I.; Soares, R.R.G.; Russom, A. Flex Printed Circuit Board Implemented Graphene-Based DNA Sensor for Detection of SARS-CoV-2. IEEE Sens. J. 2021, 21, 13060–13067. [Google Scholar] [CrossRef]
- Ji, D.; Guo, M.; Wu, Y.; Liu, W.; Luo, S.; Wang, X.; Kang, H.; Chen, Y.; Dai, C.; Kong, D.; et al. Electrochemical Detection of a Few Copies of Unamplified SARS-CoV-2 Nucleic Acids by a Self-Actuated Molecular System. J. Am. Chem. Soc. 2022, 144, 13526–13537. [Google Scholar] [CrossRef]
- Silva, L.R.G.; Stefano, J.S.; Orzari, L.O.; Brazaca, L.C.; Carrilho, E.; Marcolino-Junior, L.H.; Bergamini, M.F.; Munoz, R.A.A.; Janegitz, B.C. Electrochemical Biosensor for SARS-CoV-2 CDNA Detection Using AuPs-Modified 3D-Printed Graphene Electrodes. Biosensors 2022, 12, 622. [Google Scholar] [CrossRef]
- Amouzadeh Tabrizi, M.; Acedo, P. An Electrochemical Membrane-Based Aptasensor for Detection of Severe Acute Respiratory Syndrome Coronavirus-2 Receptor-Binding Domain. Appl. Surf. Sci. 2022, 598, 153867. [Google Scholar] [CrossRef]
- Amouzadeh Tabrizi, M.; Acedo, P. Highly Sensitive Aptasensor for the Detection of SARS-CoV-2-RBD Using Aptamer-Gated Methylene Blue@mesoporous Silica Film/Laser Engraved Graphene Electrode. Biosens. Bioelectron. 2022, 215, 114556. [Google Scholar] [CrossRef]
- Primpray, V.; Kamsong, W.; Pakapongpan, S.; Phochakum, K.; Kaewchaem, A.; Sappat, A.; Wisitsoraat, A.; Lomas, T.; Tuantranont, A.; Karuwan, C. An Alternative Ready-to-Use Electrochemical Immunosensor for Point-of-Care COVID-19 Diagnosis Using Graphene Screen-Printed Electrodes Coupled with a 3D-Printed Portable Potentiostat. Talanta Open. 2022, 6, 100155. [Google Scholar] [CrossRef]
- Kumar, T.H.V.; Srinivasan, S.; Krishnan, V.; Vaidyanathan, R.; Babu, K.A.; Natarajan, S.; Veerapandian, M. Peptide-Based Direct Electrochemical Detection of Receptor Binding Domains of SARS-CoV-2 Spike Protein in Pristine Samples. Sens. Actuators B Chem. 2023, 377, 133052. [Google Scholar] [CrossRef] [PubMed]
- Liv, L.; Çoban, G.; Nakiboğlu, N.; Kocagöz, T. A Rapid, Ultrasensitive Voltammetric Biosensor for Determining SARS-CoV-2 Spike Protein in Real Samples. Biosens. Bioelectron. 2021, 192, 113497. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, S.A.; Golab Behbahan, N.G.; Bahrani, S.; Mousavi, S.M.; Gholami, A.; Ramakrishna, S.; Firoozsani, M.; Moghadami, M.; Lankarani, K.B.; Omidifar, N. Ultra-Sensitive Viral Glycoprotein Detection NanoSystem toward Accurate Tracing SARS-CoV-2 in Biological/Non-Biological Media. Biosens. Bioelectron. 2021, 171, 112731. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Liu, F.; Xie, W.; Zhou, T.-C.; OuYang, J.; Jin, L.; Li, H.; Zhao, C.-Y.; Zhang, L.; Wei, J.; et al. Ultrasensitive Supersandwich-Type Electrochemical Sensor for SARS-CoV-2 from the Infected COVID-19 Patients Using a Smartphone. Sens. Actuators B Chem. 2021, 327, 128899. [Google Scholar] [CrossRef]
- Sadique, M.A.; Yadav, S.; Ranjan, P.; Khan, R.; Khan, F.; Kumar, A.; Biswas, D. Highly Sensitive Electrochemical Immunosensor Platforms for Dual Detection of SARS-CoV-2 Antigen and Antibody Based on Gold Nanoparticle Functionalized Graphene Oxide Nanocomposites. ACS Appl. Bio. Mater. 2022, 5, 2421–2430. [Google Scholar] [CrossRef]
- Yakoh, A.; Pimpitak, U.; Rengpipat, S.; Hirankarn, N.; Chailapakul, O.; Chaiyo, S. Paper-Based Electrochemical Biosensor for Diagnosing COVID-19: Detection of SARS-CoV-2 Antibodies and Antigen. Biosens. Bioelectron. 2021, 176, 112912. [Google Scholar] [CrossRef]
- Liv, L.; Baş, A. Discriminative Electrochemical Biosensing of Wildtype and Omicron Variant of SARS-CoV-2 Nucleocapsid Protein with Single Platform. Anal. Biochem. 2022, 657, 114898. [Google Scholar] [CrossRef]
- El-Said, W.A.; Al-Bogami, A.S.; Alshitari, W. Synthesis of Gold Nanoparticles@reduced Porous Graphene-Modified ITO Electrode for Spectroelectrochemical Detection of SARS-CoV-2 Spike Protein. Spectrochim. Acta Part. A Mol. Biomol. Spectrosc. 2022, 264, 120237. [Google Scholar] [CrossRef]
- Braz, B.A.; Hospinal-Santiani, M.; Martins, G.; Pinto, C.S.; Zarbin, A.J.G.; Beirão, B.C.B.; Thomaz-Soccol, V.; Bergamini, M.F.; Marcolino-Junior, L.H.; Soccol, C.R. Graphene-Binding Peptide in Fusion with SARS-CoV-2 Antigen for Electrochemical Immunosensor Construction. Biosensors 2022, 12, 885. [Google Scholar] [CrossRef]
- Martins, G.; Gogola, J.L.; Budni, L.H.; Papi, M.A.; Bom, M.A.T.; Budel, M.L.T.; de Souza, E.M.; Müller-Santos, M.; Beirão, B.C.B.; Banks, C.E.; et al. Novel Approach Based on GQD-PHB as Anchoring Platform for the Development of SARS-CoV-2 Electrochemical Immunosensor. Anal. Chim. Acta 2022, 1232, 340442. [Google Scholar] [CrossRef]
- Yunus, S.; Jonas, A.M.; Lakard, B. Potentiometric Biosensors. In Encyclopedia of Biophysics; Roberts, G.C.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 1941–1946. ISBN 978-3-642-16712-6. [Google Scholar]
- Mu, L.; Chang, Y.; Sawtelle, S.D.; Wipf, M.; Duan, X.; Reed, M.A. Silicon Nanowire Field-Effect Transistors—A Versatile Class of Potentiometric Nanobiosensors. IEEE Access 2015, 3, 287–302. [Google Scholar] [CrossRef]
- Alireza Hashemi, S.; Bahrani, S.; Mojtaba Mousavi, S.; Omidifar, N.; Ghaleh Golab Behbahan, N.; Arjmand, M.; Ramakrishna, S.; Bagheri Lankarani, K.; Moghadami, M.; Shokripour, M.; et al. Ultra-Precise Label-Free Nanosensor Based on Integrated Graphene with Au Nanostars toward Direct Detection of IgG Antibodies of SARS-CoV-2 in Blood. J. Electroanal. Chem. 2021, 894, 115341. [Google Scholar] [CrossRef]
- Li, Y.; Peng, Z.; Holl, N.J.; Hassan, M.R.; Pappas, J.M.; Wei, C.; Izadi, O.H.; Wang, Y.; Dong, X.; Wang, C.; et al. MXene–Graphene Field-Effect Transistor Sensing of Influenza Virus and SARS-CoV-2. ACS Omega 2021, 6, 6643–6653. [Google Scholar] [CrossRef] [PubMed]
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.-H.; Choi, M.; Ku, K.B.; Lee, C.-S.; Jun, S.; Park, D.; Kim, H.G.; et al. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142. [Google Scholar] [CrossRef] [PubMed]
- Novodchuk, I.; Kayaharman, M.; Prassas, I.; Soosaipillai, A.; Karimi, R.; Goldthorpe, I.A.; Abdel-Rahman, E.; Sanderson, J.; Diamandis, E.P.; Bajcsy, M.; et al. Electronic Field Effect Detection of SARS-CoV-2 N-Protein before the Onset of Symptoms. Biosens. Bioelectron. 2022, 210, 114331. [Google Scholar] [CrossRef] [PubMed]
- Wasfi, A.; Awwad, F.; Qamhieh, N.; Al Murshidi, B.; Palakkott, A.R.; Gelovani, J.G. Real-Time COVID-19 Detection via Graphite Oxide-Based Field-Effect Transistor Biosensors Decorated with Pt/Pd Nanoparticles. Sci. Rep. 2022, 12, 18155. [Google Scholar] [CrossRef]
- Krsihna, B.V.; Ahmadsaidulu, S.; Teja, S.S.T.; Jayanthi, D.; Navaneetha, A.; Reddy, P.R.; Prakash, M.D. Design and Development of Graphene FET Biosensor for the Detection of SARS-CoV-2. Silicon 2022, 14, 5913–5921. [Google Scholar] [CrossRef]
- Li, J.; Wu, D.; Yu, Y.; Li, T.; Li, K.; Xiao, M.-M.; Li, Y.; Zhang, Z.-Y.; Zhang, G.-J. Rapid and Unamplified Identification of COVID-19 with Morpholino-Modified Graphene Field-Effect Transistor Nanosensor. Biosens. Bioelectron. 2021, 183, 113206. [Google Scholar] [CrossRef]
- Jang, H.-J.; Sui, X.; Zhuang, W.; Huang, X.; Chen, M.; Cai, X.; Wang, Y.; Ryu, B.; Pu, H.; Ankenbruck, N.; et al. Remote Floating-Gate Field-Effect Transistor with 2-Dimensional Reduced Graphene Oxide Sensing Layer for Reliable Detection of SARS-CoV-2 Spike Proteins. ACS Appl. Mater. Interfaces 2022, 14, 24187–24196. [Google Scholar] [CrossRef]
- Chai, C.; Oh, S.-W. Electrochemical Impedimetric Biosensors for Food Safety. Food Sci. Biotechnol. 2020, 29, 879–887. [Google Scholar] [CrossRef]
- Kim, S.M.; Kim, J.; Yim, G.; Ahn, H.J.; Lee, M.; Kim, T.-H.; Park, C.; Min, J.; Jang, H.; Lee, T. Fabrication of a Surface-Enhanced Raman Spectroscopy-Based Analytical Method Consisting of Multifunctional DNA Three-Way Junction-Conjugated Porous Gold Nanoparticles and Au-Te Nanoworm for C-Reactive Protein Detection. Anal. Bioanal. Chem. 2022, 414, 3197–3204. [Google Scholar] [CrossRef] [PubMed]
- Leva-Bueno, J.; Peyman, S.A.; Millner, P.A. A Review on Impedimetric Immunosensors for Pathogen and Biomarker Detection. Med. Microbiol. Immunol. 2020, 209, 343–362. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, J.; Pumera, M. 3D-Printed COVID-19 Immunosensors with Electronic Readout. Chem. Eng. J. 2021, 425, 131433. [Google Scholar] [CrossRef]
- Ehsan, M.A.; Khan, S.A.; Rehman, A. Screen-Printed Graphene/Carbon Electrodes on Paper Substrates as Impedance Sensors for Detection of Coronavirus in Nasopharyngeal Fluid Samples. Diagnostics 2021, 11, 1030. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.; Zhang, G.F.; Hu, C.; Yuan, B.; Jahan, S.; Kitsios, G.D.; Morris, A.; Gao, S.-J.; Panat, R. Ultrarapid and Ultrasensitive Detection of SARS-CoV-2 Antibodies in COVID-19 Patients via a 3D-Printed Nanomaterial-Based Biosensing Platform. J. Med. Virol. 2022, 94, 5808–5826. [Google Scholar] [CrossRef]
- Alafeef, M.; Dighe, K.; Moitra, P.; Pan, D. Rapid, Ultrasensitive, and Quantitative Detection of SARS-CoV-2 Using Antisense Oligonucleotides Directed Electrochemical Biosensor Chip. ACS Nano 2020, 14, 17028–17045. [Google Scholar] [CrossRef]
- Pola, C.C.; Rangnekar, S.V.; Sheets, R.; Szydłowska, B.M.; Downing, J.R.; Parate, K.W.; Wallace, S.G.; Tsai, D.; Hersam, M.C.; Gomes, C.L.; et al. Aerosol-Jet-Printed Graphene Electrochemical Immunosensors for Rapid and Label-Free Detection of SARS-CoV-2 in Saliva. 2D Mater. 2022, 9, 035016. [Google Scholar] [CrossRef]
- Ang, W.L.; Lim, R.R.X.; Ambrosi, A.; Bonanni, A. Rapid Electrochemical Detection of COVID-19 Genomic Sequence with Dual-Function Graphene Nanocolloids Based Biosensor. FlatChem 2022, 32, 100336. [Google Scholar] [CrossRef]
- Ali, M.A.; Hu, C.; Jahan, S.; Yuan, B.; Saleh, M.S.; Ju, E.; Gao, S.-J.; Panat, R. Sensing of COVID-19 Antibodies in Seconds via Aerosol Jet Nanoprinted Reduced-Graphene-Oxide-Coated 3D Electrodes. Adv. Mater. 2021, 33, 2006647. [Google Scholar] [CrossRef]
- Zaccariotto, G.C.; Silva, M.K.L.; Rocha, G.S.; Cesarino, I. A Novel Method for the Detection of SARS-CoV-2 Based on Graphene-Impedimetric Immunosensor. Materials 2021, 14, 4230. [Google Scholar] [CrossRef]
- Haghayegh, F.; Salahandish, R.; Hassani, M.; Sanati-Nezhad, A. Highly Stable Buffer-Based Zinc Oxide/Reduced Graphene Oxide Nanosurface Chemistry for Rapid Immunosensing of SARS-CoV-2 Antigens. ACS Appl. Mater. Interfaces 2022, 14, 10844–10855. [Google Scholar] [CrossRef] [PubMed]
- Greben, K.; Kovalchuk, S.; Valencia, A.M.; Kirchhof, J.N.; Heeg, S.; Rietsch, P.; Reich, S.; Cocchi, C.; Eigler, S.; Bolotin, K.I. In Situ Functionalization of Graphene. 2D Mater. 2020, 8, 015022. [Google Scholar] [CrossRef]
- Schirowski, M.; Hauke, F.; Hirsch, A. Controlling the Degree of Functionalization: In-Depth Quantification and Side-Product Analysis of Diazonium Chemistry on SWCNTs. Chem. A Eur. J. 2019, 25, 12761–12768. [Google Scholar] [CrossRef] [PubMed]
- Devasena, T.; Francis, A.P.; Ramaprabhu, S. Toxicity of Graphene: An Update. In Reviews of Environmental Contamination and Toxicology; de Voogt, P., Ed.; Reviews of Environmental Contamination and Toxicology; Springer International Publishing: Cham, Switzerland, 2021; Volume 259, pp. 51–76. ISBN 978-3-030-88342-3. [Google Scholar]
- Liao, C.; Li, Y.; Tjong, S.C. Graphene Nanomaterials: Synthesis, Biocompatibility, and Cytotoxicity. Int. J. Mol. Sci. 2018, 19, 3564. [Google Scholar] [CrossRef] [PubMed]
- Ou, L.; Song, B.; Liang, H.; Liu, J.; Feng, X.; Deng, B.; Sun, T.; Shao, L. Toxicity of Graphene-Family Nanoparticles: A General Review of the Origins and Mechanisms. Part. Fibre Toxicol. 2016, 13, 57. [Google Scholar] [CrossRef]
- Achawi, S.; Feneon, B.; Pourchez, J.; Forest, V. Structure–Activity Relationship of Graphene-Based Materials: Impact of the Surface Chemistry, Surface Specific Area and Lateral Size on Their In Vitro Toxicity. Nanomaterials 2021, 11, 2963. [Google Scholar] [CrossRef]
- Huang, Y.-W.; Cambre, M.; Lee, H.-J. The Toxicity of Nanoparticles Depends on Multiple Molecular and Physicochemical Mechanisms. Int. J. Mol. Sci. 2017, 18, 2702. [Google Scholar] [CrossRef]
- Frontiñan-Rubio, J.; González, V.J.; Vázquez, E.; Durán-Prado, M. Rapid and Efficient Testing of the Toxicity of Graphene-Related Materials in Primary Human Lung Cells. Sci. Rep. 2022, 12, 7664. [Google Scholar] [CrossRef]
- Fadeel, B.; Bussy, C.; Merino, S.; Vázquez, E.; Flahaut, E.; Mouchet, F.; Evariste, L.; Gauthier, L.; Koivisto, A.J.; Vogel, U.; et al. Safety Assessment of Graphene-Based Materials: Focus on Human Health and the Environment. ACS Nano 2018, 12, 10582–10620. [Google Scholar] [CrossRef]
- Adhikari, A.; Sengupta, J. Toxicity of Carbon Nanomaterials. In Environmental Applications of Carbon. Nanomaterials-Based Devices; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2021; pp. 147–171. ISBN 978-3-527-83097-8. [Google Scholar]
- Sengupta, J.; Hussain, C.M. Prospective Pathways of Green Graphene-Based Lab-on-Chip Devices: The Pursuit toward Sustainability. Microchim. Acta 2022, 189, 177. [Google Scholar] [CrossRef]
- El Messaoudi, N.; El Mouden, A.; Fernine, Y.; El Khomri, M.; Bouich, A.; Faska, N.; Ciğeroğlu, Z.; Américo-Pinheiro, J.H.P.; Jada, A.; Lacherai, A. Green Synthesis of Ag2O Nanoparticles Using Punica Granatum Leaf Extract for Sulfamethoxazole Antibiotic Adsorption: Characterization, Experimental Study, Modeling, and DFT Calculation. Environ. Sci. Pollut. Res. 2022. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Wang, X.; Li, Z.; Zhang, S.; Xing, F. Recent Advances in the Fabrication and Application of Graphene Microfluidic Sensors. Micromachines 2020, 11, 1059. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, S.; Mathew, M.; Sekhar Rout, C. Microfluidic Sensors Based on Two-Dimensional Materials for Chemical and Biological Assessments. Mater. Adv. 2022, 3, 1874–1904. [Google Scholar] [CrossRef]
- Wang, X.; Walt, D.R. Simultaneous Detection of Small Molecules, Proteins and MicroRNAs Using Single Molecule Arrays. Chem. Sci. 2020, 11, 7896–7903. [Google Scholar] [CrossRef] [PubMed]
- Murray, L.P.; Baillargeon, K.R.; Bricknell, J.R.; Mace, C.R. Determination of Sample Stability for Whole Blood Parameters Using Formal Experimental Design. Anal. Methods 2019, 11, 930–935. [Google Scholar] [CrossRef]
- Perez, V.P.; Pessoa, W.F.B.; Galvão, B.H.A.; Sousa, E.S.S.; Dejani, N.N.; Campana, E.H.; Cavalcanti, M.G.d.S.; Cantarelli, V.V. Evaluation of Alternative RNA Extraction Methods for Detection of SARS-CoV-2 in Nasopharyngeal Samples Using the Recommended CDC Primer-Probe Set. J. Clin. Virol. Plus 2021, 1, 100032. [Google Scholar] [CrossRef]
- de Lima, L.F.; Ferreira, A.L.; Torres, M.D.T.; de Araujo, W.R.; de la Fuente-Nunez, C. Minute-Scale Detection of SARS-CoV-2 Using a Low-Cost Biosensor Composed of Pencil Graphite Electrodes. Proc. Natl. Acad. Sci. USA 2021, 118, e2106724118. [Google Scholar] [CrossRef]
- Islam, A.; Mukherjee, B.; Pandey, K.K.; Keshri, A.K. Ultra-Fast, Chemical-Free, Mass Production of High Quality Exfoliated Graphene. ACS Nano 2021, 15, 1775–1784. [Google Scholar] [CrossRef]
- Sun, Z.; Hu, Y.H. Ultrafast, Low-Cost, and Mass Production of High-Quality Graphene. Angew. Chem. Int. Ed. 2020, 59, 9232–9234. [Google Scholar] [CrossRef]
- Mbayachi, V.B.; Ndayiragije, E.; Sammani, T.; Taj, S.; Mbuta, E.R.; Khan, A.U. Graphene Synthesis, Characterization and Its Applications: A Review. Results Chem. 2021, 3, 100163. [Google Scholar] [CrossRef]
- Hui, F.; Vajha, P.; Ji, Y.; Pan, C.; Grustan-Gutierrez, E.; Duan, H.; He, P.; Ding, G.; Shi, Y.; Lanza, M. Variability of Graphene Devices Fabricated Using Graphene Inks: Atomic Force Microscope Tips. Surf. Coat. Technol. 2017, 320, 391–395. [Google Scholar] [CrossRef]
- Lebedev, A.A.; Davydov, V.Y.; Usachov, D.Y.; Lebedev, S.P.; Smirnov, A.N.; Levitskii, V.S.; Eliseyev, I.A.; Alekseev, P.A.; Dunaevskiy, M.S.; Rybkin, A.G.; et al. Study of Properties and Development of Sensors Based on Graphene Films Grown on SiC (0001) by Thermal Destruction Method. J. Phys. Conf. Ser. 2018, 951, 012007. [Google Scholar] [CrossRef]
- Zhu, Y.; Qu, B.; Andreeva, D.V.; Ye, C.; Novoselov, K.S. Graphene Standardization: The Lesson from the East. Mater. Today 2021, 47, 9–15. [Google Scholar] [CrossRef]
- Mourdikoudis, S.; Pallares, R.M.; Thanh, N.T.K. Characterization Techniques for Nanoparticles: Comparison and Complementarity upon Studying Nanoparticle Properties. Nanoscale 2018, 10, 12871–12934. [Google Scholar] [CrossRef]
- Sadique, M.A.; Ranjan, P.; Yadav, S.; Khan, R. Chapter 8—Advanced High-Throughput Biosensor-Based Diagnostic Approaches for Detection of Severe Acute Respiratory Syndrome-Coronavirus-2. In Computational Approaches for Novel Therapeutic and Diagnostic Designing to Mitigate SARS-CoV-2 Infection; Parihar, A., Khan, R., Kumar, A., Kaushik, A.K., Gohel, H., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 147–169. ISBN 978-0-323-91172-6. [Google Scholar]
- Venugopalan, A. COVID-19: IIT Kharagpur Researchers Come up with Portable, Low Cost and Fast Testing Device. Economic Times, 25 July 2020. [Google Scholar]
- Chauhan, N.; Maekawa, T.; Kumar, D.N.S. Graphene Based Biosensors—Accelerating Medical Diagnostics to New-Dimensions. J. Mater. Res. 2017, 32, 2860–2882. [Google Scholar] [CrossRef]
- Center for Devices and Radiological Health In Vitro Diagnostics EUAs—Antigen Diagnostic Tests for SARS-CoV-2. Available online: https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas-antigen-diagnostic-tests-sars-cov-2 (accessed on 11 April 2023).
- Diagnostic Kit Evaluation. Available online: https://www.icmr.gov.in/ckitevaluation.html (accessed on 11 April 2023).
- Teo, A.K.J.; Choudhury, Y.; Tan, I.B.; Cher, C.Y.; Chew, S.H.; Wan, Z.Y.; Cheng, L.T.E.; Oon, L.L.E.; Tan, M.H.; Chan, K.S.; et al. Saliva Is More Sensitive than Nasopharyngeal or Nasal Swabs for Diagnosis of Asymptomatic and Mild COVID-19 Infection. Sci. Rep. 2021, 11, 3134. [Google Scholar] [CrossRef]
- Byrne, R.L.; Kay, G.A.; Kontogianni, K.; Aljayyoussi, G.; Brown, L.; Collins, A.M.; Cuevas, L.E.; Ferreira, D.M.; Fraser, A.J.; Garrod, G.; et al. Saliva Alternative to Upper Respiratory Swabs for SARS-CoV-2 Diagnosis. Emerg. Infect. Dis. J. CDC 2020, 26, 2769–2770. [Google Scholar] [CrossRef]
- Labbé, A.-C.; Benoit, P.; Gobeille Paré, S.; Coutlée, F.; Lévesque, S.; Bestman-Smith, J.; Dumaresq, J.; Lavallée, C.; Houle, C.; Martin, P.; et al. Comparison of Saliva with Oral and Nasopharyngeal Swabs for SARS-CoV-2 Detection on Various Commercial and Laboratory-Developed Assays. J. Med. Virol. 2021, 93, 5333–5338. [Google Scholar] [CrossRef]
- Bastos, M.L.; Perlman-Arrow, S.; Menzies, D.; Campbell, J.R. The Sensitivity and Costs of Testing for SARS-CoV-2 Infection With Saliva Versus Nasopharyngeal Swabs. Ann. Intern. Med. 2021, 174, 501–510. [Google Scholar] [CrossRef]
- Angcard COVID-19 Saliva Home Testing Kits. Available online: https://www.angstrombiotech.in/angcard.html (accessed on 11 April 2023).
- Vogels, C.B.F.; Watkins, A.E.; Harden, C.A.; Brackney, D.E.; Shafer, J.; Wang, J.; Caraballo, C.; Kalinich, C.C.; Ott, I.M.; Fauver, J.R.; et al. SalivaDirect: A Simplified and Flexible Platform to Enhance SARS-CoV-2 Testing Capacity. Med 2021, 2, 263–280.e6. [Google Scholar] [CrossRef]
- Xie, J.-W.; He, Y.; Zheng, Y.-W.; Wang, M.; Lin, Y.; Lin, L.-R. Diagnostic Accuracy of Rapid Antigen Test for SARS-CoV-2: A Systematic Review and Meta-analysis of 166,943 Suspected COVID-19 Patients. Microbiol. Res. 2022, 265, 127185. [Google Scholar] [CrossRef] [PubMed]
- Olearo, F.; Nörz, D.; Heinrich, F.; Sutter, J.P.; Roedl, K.; Schultze, A.; Schulze zur Wiesch, J.; Braun, P.; Oestereich, L.; Kreuels, B.; et al. Handling and Accuracy of Four Rapid Antigen Tests for the Diagnosis of SARS-CoV-2 Compared to RT-QPCR. J. Clin. Virol. 2021, 137, 104782. [Google Scholar] [CrossRef]
- Jegerlehner, S.; Suter-Riniker, F.; Jent, P.; Bittel, P.; Nagler, M. Diagnostic Accuracy of a SARS-CoV-2 Rapid Antigen Test in Real-Life Clinical Settings. Int. J. Infect. Dis. 2021, 109, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Arshadi, M.; Fardsanei, F.; Deihim, B.; Farshadzadeh, Z.; Nikkhahi, F.; Khalili, F.; Sotgiu, G.; Shahidi Bonjar, A.H.; Centis, R.; Migliori, G.B.; et al. Diagnostic Accuracy of Rapid Antigen Tests for COVID-19 Detection: A Systematic Review With Meta-Analysis. Front. Med. 2022, 9, 870738. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Poudel, A.; Karki, D.; Thapa, J. Diagnostic Accuracy of Antigen-Detection Rapid Diagnostic Tests for Diagnosis of COVID-19 in Low-and Middle-Income Countries: A Systematic Review and Meta-Analysis. PLoS Glob. Public Health 2022, 2, e0000358. [Google Scholar] [CrossRef] [PubMed]
- Khandker, S.S.; Nik Hashim, N.H.H.; Deris, Z.Z.; Shueb, R.H.; Islam, M.A. Diagnostic Accuracy of Rapid Antigen Test Kits for Detecting SARS-CoV-2: A Systematic Review and Meta-Analysis of 17,171 Suspected COVID-19 Patients. J. Clin. Med. 2021, 10, 3493. [Google Scholar] [CrossRef]
- Dinnes, J.; Sharma, P.; Berhane, S.; Wyk, S.S.; Nyaaba, N.; Domen, J.; Taylor, M.; Cunningham, J.; Davenport, C.; Dittrich, S.; et al. Rapid, Point-of-care Antigen Tests for Diagnosis of SARS-CoV-2 Infection. Cochrane Database Syst. Rev. 2022, 2022, CD013705. [Google Scholar] [CrossRef]














| Protein | Function |
|---|---|
| Spike (S) | Binds and fuse to the host cell receptor and induces infection and transmission. |
| Nucleocapsid (N) | Binding to the viral RNA genome is critical for viral replication and genome packaging. |
| Membrane (M) | Viral assembly and shaping viral envelope. |
| Envelope (E) | Formation of the viral envelope. |
| Biosensor Type | Pros | Cons |
|---|---|---|
| Amperometric | High sensitivity, low detection limit, and fast response time | Requires a high potential to operate, susceptible to interference from other electroactive species, may suffer from electrode fouling and drift over time, may require a redox mediator |
| Potentiometric | Low cost, easy to use, and good stability, suitable for continuous real-time monitoring, | Limited sensitivity, slow response time, large sensor size, high cost, the signal can be affected by pH and temperature changes |
| Impedimetric | High sensitivity, low detection limit, fast response time, good specificity, ability to detect changes in interfacial properties | Requires complex instrumentation, lower sensitivity compared to amperometric and potentiometric biosensors, can be affected by variations in solution conductivity, may require additional instrumentation for measurement, may require specialized surface functionalization |
| Key Carbon Material | Target | Limit of Detection (LOD) | Detection Range | Sensitivity | Response Time | Ref. |
|---|---|---|---|---|---|---|
| Graphene | Nucleocapsid protein, S1-IgG, S1-IgM and C-reactive protein | Nucleocapsid protein 0.1 μg/mL (serum) and 0.5 ng/mL (saliva) S1-IgG 2 μg/mL (serum) and 0.2 μg/mL (saliva) S1-IgM 20 μg/mL (serum) and 0.6 μg/mL (saliva) CRP 10 μg/mL (serum) and 0.1 μg/mL (saliva) | Nucleocapsid protein 0.1–0.8 μg/mL (serum) and 0.5–2.0 ng/mL (saliva) S1-IgG 2–40 μg/mL (serum) and 0.2–0.5 μg/mL (saliva) S1-IgM 20–50 μg/mL (serum) and 0.6–5.0 μg/mL (saliva) CRP 10–20 μg/mL (serum) and 0.1–0.5 μg/mL (saliva) | 16.28 nA mL ng−1 | 1 min | Rodríguez et al. [54] |
| Nucleocapsid Protein | 2.0 fg/mL | 0.1 pg/mL to 500 ng/mL | NA | 120 min (incubation time) | Jaewjaroenwattana et al. [55] | |
| Nucleocapsid Antibody | 13 fM | 100 fM to 1 nM | NA | 10 s | Ali et al. [56] | |
| Spike protein | 2.9 ng/mL | 2.9 to 500 ng/mL | NA | 60 min (incubation time) | Beduk et al. [57] | |
| Spike protein | 260 nM | NA | NA | 45 min (incubation time) | Mojsoska et al. [58] | |
| Delta Variant (RNA) | 1.2 pM | 4 pM to 4 nM | NA | 47 min | Yang et al. [59] | |
| RNA | 100 fg/mL | 100 fg/mL to 1μg/mL | NA | 30 min (hybridization time) | Damiati et al. [60] | |
| RNA | 0.025 copies/μL | NA | NA | 30 min for incubating, less than 1 min for detection | Ji et al. [61] | |
| cDNA | 0.30 µmol/L | NA | 0.583 µA µmol−1 L | 30 min (hybridization time) | Silva et al. [62] | |
| RBD | 0.8 ng/mL | 2.5 to 40.0 ng/mL | NA | 20 min (incubation time) | Tabrizi et al. [63] | |
| RBD | 0.36 ng/mL | 0.5 to 250 ng/mL | NA | 30 min | Tabrizi et al. [64] | |
| RBD | 2 pg/mL | 0.01 to 1500 ng/mL | NA | 30 min (incubation time) | Primpray et al. [65] | |
| Graphene Oxide | Spike protein | 0.58 pg/mL | 1 pg/mL to 1 µg/mL | 0.0105 mA/pg mL−1 cm−2 | NA | Kumar et al. [66] |
| Spike Protein | 1 ag/mL | 1 ag/mL to 10 fg/mL | 93.3% | 5 min | Liv et al. [67] | |
| Glycoprotein | 1.68 × 10−22 μg/mL | NA | 0.0048 μAμg·mL−1·cm−2 | 1 min | Hashemi et al. [68] | |
| RNA | 200 copies/mL | NA | NA | NA | Zhao et al. [69] | |
| Nucleocapsid protein and immunoglobulin (Ig) G | Antigen (3.99 ag/mL) Antibody (1.0 fg/mL) | Antigen (10.0 ag/mL to 50.0 ng/mL) Antibody (1.0 fg/mL to 1.0 ng/mL) | NA | NA | Sadique et al. [70] | |
| IgG and IgM | 1 ng/mL | 1 to 1000 ng/mL | 100% | 30 min (incubation time) | Yakoh et al. [71] | |
| Nucleocapsid protein | 0.24 ag/mL | 1 ag/mL to 10 fg/mL | NA | NA | Liv et al. [72] | |
| Reduced Graphene Oxide | Spike protein | 39.5 fmol/L | 100 nmol/L to 500 fmol/L | NA | NA | El-Said et al. [73] |
| Immunoglobulin G | 0.77 μg/mL | NA | NA | 60 min | Braz et al. [74] | |
| Graphene Quantum Dots | Anti-S antibodies (IgG) | 100 ng/mL | 100 ng/mL to 10 μg/mL | NA | 120 min | Martins et al. [75] |
| Key Carbon Material | Target | Limit of Detection (LOD) | Detection Range | Sensitivity | Response Time | Ref. |
|---|---|---|---|---|---|---|
| Graphene | IgG antibodies | 0.18 × 10−19% V/V | NA | 2.14 μA% V/V·cm−2 | 1 min | Hashemi et al. [78] |
| Spike Protein | 1 fg/mL | 1 fg/mL to 10 pg/mL | NA | 50 ms | Li et al. [79] | |
| Spike protein | 1 fg/mL | NA | NA | 1 min | Seo et al. [80] | |
| Graphene oxide | Nucleocapsid protein | 10 ag/mL | 10 ag/mL to 1 μg/mL | NA | 4 min | Novodchuk et al. [81] |
| Spike protein | 1 fg/mL | 1 fg/mL to 100 ng/mL) | NA | NA | Wasfi et al. [82] | |
| Reduced graphene oxide | spike protein | 0.002 fM | NA | NA | NA | Krsihna et al. [83] |
| RNA | PBS (0.37 fM), throat swab (2.29 fM), and serum (3.99 fM) | NA | NA | 2 min | Li et al. [84] | |
| Spike protein | 3.4 pg/mL | 500 fg/mL to 5 μg/mL | 5.1 mV/dec | NA | Jang et al. [85] |
| Key Carbon Material | Target | Limit of Detection (LOD) | Detection Range | Sensitivity | Response Time | Ref. |
|---|---|---|---|---|---|---|
| Graphene | Spike Protein | 0.5 ± 0.1 μg/mL | 1.0 to 10 μg/mL | 0.076 ppm−1 | 20 min | Muñoz et al. [89] |
| Spike Protein | 0.25 fg/mL | 0.25 fg/mL to 1 µg/mL | NA | 5 min | Ehsan et al. [90] | |
| Nucleocapsid (N), spike 1 (S1), and RBD proteins | 1 pm (N), 0.1 pm (S1), 10 fM (RBD) | 10 fM to 50 nM | 100% | 10–12 s | Ali et al. [91] | |
| Nucleocapsid phosphoprotein (N-gene) | 6.9 copies/μL | 585.4 to 5.854 × 107 copies/μL | 231 (copies μL−1)−1 | 5 min | Alafeef et al. [92] | |
| Spike RBD | 22.91 ± 4.72 pg/mL | 1 to 1000 ng/mL | NA | 30 min | Pola et al. [93] | |
| Graphene Oxide | Gene | 186 × 10−9 M | 10−10 to 10−5 M | NA | NA | Ang et al. [94] |
| Reduced Graphene Oxide | Spike (S1) and RBD proteins | 2.8 × 10−15 M (S1) 16.9 × 10−15 M (RBD) | NA | 1 × 10−12 M (S1) 1 × 10−15 M (RBD) | 12 s | Ali et al. [95] |
| spike protein RBD | 150 ng/mL | 0.16 to 40 μg/mL | NA | NA | Zaccariotto et al. [96] | |
| nucleocapsid (N-) protein antigens | 21 fg/mL | 1 to 10,000 pg/mL | 32.07 ohms·mL/ pg·mm2 | 15 min | Haghayegh et al. [97] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sengupta, J.; Hussain, C.M. Graphene-Based Electrochemical Nano-Biosensors for Detection of SARS-CoV-2. Inorganics 2023, 11, 197. https://doi.org/10.3390/inorganics11050197
Sengupta J, Hussain CM. Graphene-Based Electrochemical Nano-Biosensors for Detection of SARS-CoV-2. Inorganics. 2023; 11(5):197. https://doi.org/10.3390/inorganics11050197
Chicago/Turabian StyleSengupta, Joydip, and Chaudhery Mustansar Hussain. 2023. "Graphene-Based Electrochemical Nano-Biosensors for Detection of SARS-CoV-2" Inorganics 11, no. 5: 197. https://doi.org/10.3390/inorganics11050197
APA StyleSengupta, J., & Hussain, C. M. (2023). Graphene-Based Electrochemical Nano-Biosensors for Detection of SARS-CoV-2. Inorganics, 11(5), 197. https://doi.org/10.3390/inorganics11050197