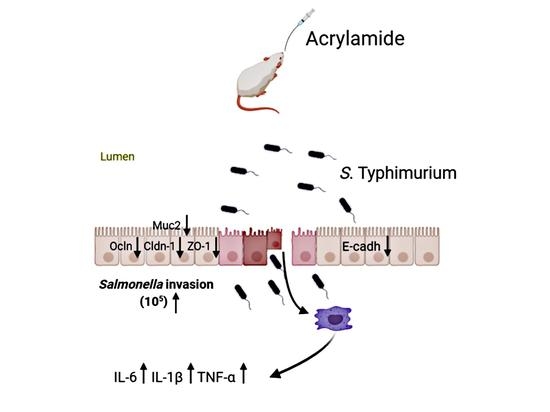
Dietary Acrylamide Intake Alters Gut Microbiota in Mice and Increases Its Susceptibility to Salmonella Typhimurium Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Animals Treatment
2.3. S. Typhimurium Infection
2.4. Faecal Samples and Microbiota Profiling
2.5. Histological Analyses
2.6. RNA Extraction and RT-qPCR Analysis
2.7. Bacterial CFU Determination
2.8. Western Blot Analysis
2.9. Statistic Analysis
3. Results
3.1. The Effect of AA on the Body Weight, Food and Water Intake
3.2. The Effect of AA on the Diversity and Composition of Gut Microbiota
3.3. The Effect of AA on Host Susceptibility to S. Typhimurium Infection
3.4. The Effect of AA on Inflammatory Cytokines
3.5. The Effect of AA on Intestine Epithelial Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Kreuzer, M.; Hardt, W.-D. How Food Affects Colonization Resistance Against Enteropathogenic Bacteria. Annu. Rev. Microbiol. 2020, 74, 787–813. [Google Scholar] [CrossRef]
- Buffie, C.G.; Pamer, E.G. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 2013, 13, 790–801. [Google Scholar] [CrossRef] [Green Version]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Núñez, G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef]
- Doorduyn, Y.; Van Den Brandhof, W.E.; Van Duynhoven, Y.T.H.P.; Wannet, W.J.B.; Van Pelt, W. Risk factors for Salmonella Enteritidis and Typhimurium (DT104 and non-DT104) infections in The Netherlands: Predominant roles for raw eggs in Enteritidis and sandboxes in Typhimurium infections. Epidemiol. Infect. 2005, 134, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, A.; Lam, L.; Rajendram, M.; Tamburini, F.; Honeycutt, J.; Pham, T.; Van Treuren, W.; Pruss, K.; Stabler, S.R.; Lugo, K.; et al. A Gut Commensal-Produced Metabolite Mediates Colonization Resistance to Salmonella Infection. Cell Host Microbe 2018, 24, 296–307.e297. [Google Scholar] [CrossRef] [Green Version]
- Sekirov, I.; Tam, N.M.; Jogova, M.; Robertson, M.L.; Li, Y.; Lupp, C.; Finlay, B.B. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 2008, 76, 4726–4736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaudhuri, D.; Roy Chowdhury, A.; Biswas, B.; Chakravortty, D. Salmonella Typhimurium Infection Leads to Colonization of the Mouse Brain and Is Not Completely Cured With Antibiotics. Front. Microbiol. 2018, 9, 1632. [Google Scholar] [CrossRef]
- Boumart, Z.; Velge, P.; Wiedemann, A. Multiple invasion mechanisms and different intracellular Behaviors: A new vision of Salmonella–host cell interaction. FEMS Microbiol. Lett. 2014, 361, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barthel, M.; Hapfelmeier, S.; Quintanilla-Martínez, L.; Kremer, M.; Rohde, M.; Hogardt, M.; Pfeffer, K.; Rüssmann, H.; Hardt, W.D. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 2003, 71, 2839–2858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawley, T.D.; Bouley, D.M.; Hoy, Y.E.; Gerke, C.; Relman, D.A.; Monack, D.M. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect. Immun. 2008, 76, 403–416. [Google Scholar] [CrossRef] [Green Version]
- Capuano, E.; Fogliano, V. Acrylamide and 5-hydroxymethylfurfural (HMF): A review on metabolism, toxicity, occurrence in food and mitigation strategies. LWT Food Sci. Technol. 2011, 44, 793–810. [Google Scholar] [CrossRef]
- Ruden, C. Acrylamide and cancer risk—Expert risk assessments and the public debate. Food Chem. Toxicol. 2004, 42, 335–349. [Google Scholar] [CrossRef]
- Rice, J.M. The carcinogenicity of acrylamide. Mutat. Res. 2005, 580, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Duda-Chodak, A.; Wajda, L.; Tarko, T.; Sroka, P.; Satora, P. A review of the interactions between acrylamide, microorganisms and food components. Food Funct. 2016, 7, 1282–1295. [Google Scholar] [CrossRef]
- Friedman, M.; Levin, C.E. Review of methods for the reduction of dietary content and toxicity of acrylamide. J. Agric. Food Chem. 2008, 56, 6113–6140. [Google Scholar] [CrossRef] [PubMed]
- Cha, M. Enzymatic control of the acrylamide level in coffee. Eur. Food Res. Technol. 2013, 236, 567–571. [Google Scholar] [CrossRef]
- Pedreschi, F.; Kaack, K.; Granby, K. Acrylamide content and color development in fried potato strips. Food Res. Int. 2006, 39, 40–46. [Google Scholar] [CrossRef]
- Pelucchi, C.; Franceschi, S.; Levi, F.; Trichopoulos, D.; Bosetti, C.; Negri, E.; La Vecchia, C. Fried potatoes and human cancer. Int. J. Cancer 2003, 105, 558–560. [Google Scholar] [CrossRef]
- Sengul, E.; Gelen, V.; Yildirim, S.; Tekin, S.; Dag, Y. The Effects of Selenium in Acrylamide-Induced Nephrotoxicity in Rats: Roles of Oxidative Stress, Inflammation, Apoptosis, and DNA Damage. Biol. Trace Elem. Res. 2021, 199, 173–184. [Google Scholar] [CrossRef] [PubMed]
- National Toxicology Program. Toxicology and carcinogenesis studies of acrylamide (CASRN 79-06-1) in F344/N rats and B6C3F1 mice (feed and drinking water studies). Natl. Toxicol. Progr. Tech. Rep. Ser. 2012, 575, 1–234. [Google Scholar]
- Friedman, M. Chemistry, biochemistry, and safety of acrylamide. A review. J. Agric. Food Chem. 2003, 51, 4504–4526. [Google Scholar] [CrossRef] [PubMed]
- Qin, N.; Song, G.; Ren, X.; Zhang, L.; Gao, J.; Xia, X.; Zhu, B. Fish oil extracted from Coregonus peled improves obese phenotype and changes gut microbiota in a high-fat diet-induced mouse model of recurrent obesity. Food Funct. 2020, 11, 6158–6169. [Google Scholar] [CrossRef]
- Drolia, R.; Tenguria, S.; Durkes, A.C.; Turner, J.R.; Bhunia, A.K. Listeria Adhesion Protein Induces Intestinal Epithelial Barrier Dysfunction for Bacterial Translocation. Cell Host Microbe 2018, 23, 470–484.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.-F.; Wu, K.-Y.; Liou, S.-H.; Uang, S.-N.; Chen, C.-C.; Shih, W.-C.; Lee, S.-C.; Huang, C.-C.J.; Chen, M.-L. Biological monitoring for occupational acrylamide exposure from acrylamide production workers. Int. Arch. Occup. Environ. Health 2011, 84, 303–313. [Google Scholar] [CrossRef]
- Pennisi, M.; Malaguarnera, G.; Puglisi, V.; Vinciguerra, L.; Vacante, M.; Malaguarnera, M. Neurotoxicity of acrylamide in exposed workers. Int. J. Environ. Res. Public Health 2013, 10, 3843–3854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, L.; Zhang, P.; Xiang, L.; Wang, H.; Rao, P.; Wang, S. Interaction of acrylamide with micelles in French fry aqueous extracts. Food Control 2020, 110, 106974. [Google Scholar] [CrossRef]
- Collado, M.C.; Isolauri, E.; Laitinen, K.; Salminen, S. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Am. J. Clin. Nutr. 2008, 88, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Bäckhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070. [Google Scholar] [CrossRef] [Green Version]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef] [Green Version]
- Hapfelmeier, S.; Hardt, W.-D. A mouse model for S. Typhimurium-induced enterocolitis. Trends Microbiol. 2005, 13, 497–503. [Google Scholar] [CrossRef]
- Bumann, D.; Cunrath, O. Heterogeneity of Salmonella-host interactions in infected host tissues. Curr. Opin. Microbiol. 2017, 39, 57–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, R.A.; Lee, C.A. Invasion genes are not required for Salmonella enterica serovar typhimurium to breach the intestinal epithelium: Evidence that salmonella pathogenicity island 1 has alternative functions during infection. Infect. Immun. 2000, 68, 5050–5055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Euzéby, J. List of new names and new combinations previously effectively, but not validly, published. Int. J. Syst. Evol. Microbiol. 2006, 56, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Godoy, D.; Randle, G.; Simpson, A.J.; Aanensen, D.M.; Pitt, T.L.; Kinoshita, R.; Spratt, B.G. Multilocus Sequence Typing and Evolutionary Relationships among the Causative Agents of Melioidosis and Glanders, Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 2003, 41, 2068–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakon, H.; Nagai, F.; Morotomi, M.; Tanaka, R. Sutterella parvirubra sp. nov. and Megamonas funiformis sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2008, 58, 970–975. [Google Scholar] [CrossRef] [PubMed]
- Molitoris, E.; Wexler, H.M.; Finegold, S.M. Sources and antimicrobial susceptibilities of Campylobacter gracilis and Sutterella wadsworthensis. Clin. Infect. Dis. 1997, 25 (Suppl. 2), S264–S265. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol. Autism 2013, 4, 42. [Google Scholar] [CrossRef] [Green Version]
- Turnbaugh, P.J.; Bäckhed, F.; Fulton, L.; Gordon, J.I. Diet-Induced Obesity Is Linked to Marked but Reversible Alterations in the Mouse Distal Gut Microbiome. Cell Host Microbe 2008, 3, 213–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; DiBaise, J.K.; Zuccolo, A.; Kudrna, D.; Braidotti, M.; Yu, Y.; Parameswaran, P.; Crowell, M.D.; Wing, R.; Rittmann, B.E.; et al. Human gut microbiota in obesity and after gastric bypass. Proc. Natl. Acad. Sci. USA 2009, 106, 2365–2370. [Google Scholar] [CrossRef] [Green Version]
- Martínez, I.; Perdicaro, D.J.; Brown, A.W.; Hammons, S.; Carden, T.J.; Carr, T.P.; Eskridge, K.M.; Walter, J. Diet-Induced Alterations of Host Cholesterol Metabolism Are Likely To Affect the Gut Microbiota Composition in Hamsters. Appl. Environ. Microbiol. 2013, 79, 516–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaakoush, N.O. Insights into the Role of Erysipelotrichaceae in the Human Host. Front. Cell. Infect. Microbiol. 2015, 5, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Craven, M.; Egan, C.E.; Dowd, S.E.; McDonough, S.P.; Dogan, B.; Denkers, E.Y.; Bowman, D.; Scherl, E.J.; Simpson, K.W. Inflammation Drives Dysbiosis and Bacterial Invasion in Murine Models of Ileal Crohn’s Disease. PLoS ONE 2012, 7, e41594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaubeck, M.; Clavel, T.; Calasan, J.; Lagkouvardos, I.; Haange, S.B.; Jehmlich, N.; Basic, M.; Dupont, A.; Hornef, M.; Bergen, M.V.; et al. Dysbiotic gut microbiota causes transmissible Crohn’s disease-like ileitis independent of failure in antimicrobial defence. Gut 2016, 65, 225–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Feng, Y.; Xu, Y.; Wu, Q.; Han, Q.A.; Liang, X.; Yang, B.; Wang, X.; Xia, X. The anti-infective activity of punicalagin against Salmonella enterica subsp. enterica serovar typhimurium in mice. Food Funct. 2015, 6, 2357–2364. [Google Scholar] [CrossRef] [PubMed]
- Zihni, C.; Mills, C.; Matter, K.; Balda, M.S. Tight junctions: From simple barriers to multifunctional molecular gates. Nat. Rev. Mol. Cell Biol. 2016, 17, 564–580. [Google Scholar] [CrossRef]
- Zeisel, M.B.; Dhawan, P.; Baumert, T.F. Tight junction proteins in gastrointestinal and liver disease. Gut 2019, 68, 547–561. [Google Scholar] [CrossRef]
- Brandner, J.M.; Zorn-Kruppa, M.; Yoshida, T.; Moll, I.; Beck, L.A.; De Benedetto, A. Epidermal tight junctions in health and disease. Tissue Barriers 2015, 3, e974451. [Google Scholar] [CrossRef] [Green Version]
- Zeissig, S.; Bürgel, N.; Günzel, D.; Richter, J.; Mankertz, J.; Wahnschaffe, U.; Kroesen, A.J.; Zeitz, M.; Fromm, M.; Schulzke, J.D. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn’s disease. Gut 2007, 56, 61–72. [Google Scholar] [CrossRef]
- Bürgel, N.; Bojarski, C.; Mankertz, J.; Zeitz, M.; Fromm, M.; Schulzke, J.D. Mechanisms of diarrhea in collagenous colitis. Gastroenterology 2002, 123, 433–443. [Google Scholar] [CrossRef]
- Schneeberger, E.E.; Lynch, R.D. The tight junction: A multifunctional complex. Am. J. Physiol. Cell Physiol. 2004, 286, C1213–C1228. [Google Scholar] [CrossRef]
- Shin, K.; Fogg, V.C.; Margolis, B. Tight junctions and cell polarity. Annu. Rev. Cell Dev. Biol. 2006, 22, 207–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Roy, F.; Berx, G. The cell-cell adhesion molecule E-cadherin. Cell. Mol. Life Sci. CMLS 2008, 65, 3756–3788. [Google Scholar] [CrossRef] [PubMed]
- Gumbiner, B.; Stevenson, B.; Grimaldi, A. The role of the cell adhesion molecule uvomorulin in the formation and maintenance of the epithelial junctional complex. J. Cell Biol. 1988, 107, 1575–1587. [Google Scholar] [CrossRef] [Green Version]
- Nikitas, G.; Deschamps, C.; Disson, O.; Niault, T.; Cossart, P.; Lecuit, M. Transcytosis of Listeria monocytogenes across the intestinal barrier upon specific targeting of goblet cell accessible E-cadherin. J. Exp. Med. 2011, 208, 2263–2277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.Y.; He, X.L.; Puthiyakunnon, S.; Peng, L.; Li, Y.; Wu, L.S.; Peng, W.L.; Zhang, Y.; Gao, J.; Zhang, Y.Y.; et al. Mucin2 is Required for Probiotic Agents-Mediated Blocking Effects on Meningitic E. coli-Induced Pathogenicities. J. Microbiol. Biotechnol. 2015, 25, 1751–1760. [Google Scholar] [CrossRef] [PubMed]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Liu, H.; Liu, J.; Ren, X.; Song, G.; Xia, X.; Qin, N. Dietary Acrylamide Intake Alters Gut Microbiota in Mice and Increases Its Susceptibility to Salmonella Typhimurium Infection. Foods 2021, 10, 2990. https://doi.org/10.3390/foods10122990
Wang Z, Liu H, Liu J, Ren X, Song G, Xia X, Qin N. Dietary Acrylamide Intake Alters Gut Microbiota in Mice and Increases Its Susceptibility to Salmonella Typhimurium Infection. Foods. 2021; 10(12):2990. https://doi.org/10.3390/foods10122990
Chicago/Turabian StyleWang, Zhen, Hongxu Liu, Jiaxiu Liu, Xiaomeng Ren, Guoku Song, Xiaodong Xia, and Ningbo Qin. 2021. "Dietary Acrylamide Intake Alters Gut Microbiota in Mice and Increases Its Susceptibility to Salmonella Typhimurium Infection" Foods 10, no. 12: 2990. https://doi.org/10.3390/foods10122990
APA StyleWang, Z., Liu, H., Liu, J., Ren, X., Song, G., Xia, X., & Qin, N. (2021). Dietary Acrylamide Intake Alters Gut Microbiota in Mice and Increases Its Susceptibility to Salmonella Typhimurium Infection. Foods, 10(12), 2990. https://doi.org/10.3390/foods10122990