Stepwise Approach for Tracing the Geographical Origins of the Manila Clam Ruditapes philippinarum Using Dual-Element Isotopes and Carbon Isotopes of Fatty Acids
Abstract
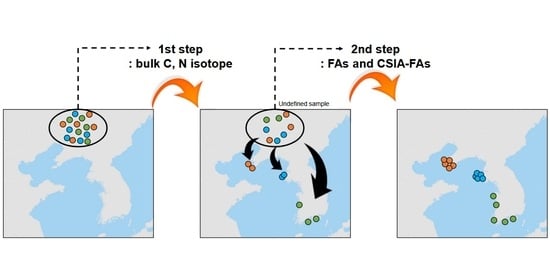
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Stable Isotope Analysis of Bulk Samples
2.3. FA Content and CSIA of Phospholipid FA
2.4. Statistical Analysis: Feature Scaling and LDA
3. Results and Discussions
3.1. First Step: A Dual-Isotope Analysis for Distinct Geographical Origins of Clams from Four Different Provinces (Coasts) of Korea
3.2. Second Step I: FA Composition for Geographical Authentication
3.3. Second Step II: Geographical Authentication Using CSIA of Carbon Isotopes in Fas
3.4. Applications for Imported Clams: Geographical Authentication Using Carbon Isotopes of FAs from Small Amounts of Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Luo, D.; Dong, H.; Luo, H.; Xian, Y.; Wan, J.; Guo, X.; Wu, Y. The application of stable isotope ratio analysis to determine the geographical origin of wheat. Food Chem. 2015, 174, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Elgadi, S.; Ouhammou, A.; Taous, F.; Zine, H.; Papazoglou, E.G.; Elghali, T.; Amenzou, N.; El Allali, H.; Aitlhaj, A.; El Antari, A. Combination of stable isotopes and fatty acid composition for geographical origin discrimination of one argan oil vintage. Foods 2021, 10, 1274. [Google Scholar] [CrossRef] [PubMed]
- European Commission. Available online: https://knowledge4policy.ec.europa.eu/food-fraud-quality_en (accessed on 27 June 2022).
- Fox, M.; Mitchell, M.; Dean, M.; Elliott, C.; Campbell, K. The seafood supply chain from a fraudulent perspective. Food Secur. 2018, 10, 939–963. [Google Scholar] [CrossRef] [Green Version]
- Asche, F.; Bjørndal, T.; Young, J.A. Market interactions for aquaculture products. Aquac. Econ. Manag. 2001, 5, 303–318. [Google Scholar] [CrossRef]
- Gopi, K.; Mazumder, D.; Sammut, J.; Saintilan, N. Determining the provenance and authenticity of seafood: A review of current methodologies. Trends. Food Sci. Technol. 2019, 91, 294–304. [Google Scholar] [CrossRef]
- Megdal, P.A.; Craft, N.A.; Handelman, G.J. A simplified method to distinguish farmed (Salmo salar) from wild salmon: Fatty acid ratios versus astaxanthic chiral isomers. Lipids 2009, 44, 569–576. [Google Scholar] [CrossRef] [Green Version]
- KIMST. Development of Practical Technique to Establish Fisheries Forensic Center. Korea Institute of Marine Science & Technology Promotion. 2017. Available online: https://scienceon.kisti.re.kr/srch/selectPORSrchReport.do?cn=TRKO201900000122 (accessed on 27 June 2022).
- Lee, D.; Seo, S.; Song, M.K.; Lee, H.K.; Park, S.; Jin, Y.W. Factors associated with the risk perception and purchase decisions of Fukushima-related food in South Korea. PLoS ONE 2017, 12, e018765. [Google Scholar] [CrossRef] [Green Version]
- Tan, Y.; Fang, L.; Qiu, M.; Huo, Z.; Yan, X. Population genetics of the manila clam (Ruditapes philippinarum) in East Asia. Sci. Rep. 2020, 10, 21890. [Google Scholar] [CrossRef]
- FAO. Food and Agriculture Organization of the United Nation FAO Yearbook. In Fishery and Aquaculture Statistics 2011; Food and Agriculture Organization of the United Nations: Rome, Italy, 2003; p. 76. [Google Scholar]
- Kim, E.; Kim, H.; Shin, K.H.; Kim, M.S.; Kundu, S.R.; Lee, B.G.; Han, S. Biomagnification of mercury through the benthic food webs of a temperate estuary: Masan Bay, Korea. Environ. Toxicol. Chem. 2012, 31, 1254–1263. [Google Scholar] [CrossRef]
- Greene, J.L. Country-of-origin labeling for foods and the WTO trade dispute on meat labeling. In Congressional Research Service (CRS) Reports; Congressional Research Service: Washington, DC, USA, 2015. [Google Scholar]
- Galstyan, A.G.; Semipyatniy, V.K.; Mikhailova, I.Y.; Gilmanov, K.K.H.; Bigaeva, A.V.; Vafin, R.R. Methodological approaches to DNA authentication of foods, wines and raw materials for their production. Foods 2021, 10, 595. [Google Scholar] [CrossRef]
- Selamat, J.; Rozani, N.A.A.; Murugesu, S. Application of the metabolomics approach in food authentication. Molecules 2021, 26, 7565. [Google Scholar] [CrossRef]
- Peterson, B.J.; Fry, B. Stable isotopes in ecosystem studies. Annu. Rev. Ecol. Syst. 1987, 18, 293–320. [Google Scholar] [CrossRef]
- Vander Zanden, M.J.; Rasmussen, J.B. Variation in δ15N and δ13C trophic fractionation: Implications for aquatic food web studies. Limnol. Oceanogr. 2001, 46, 2061–2066. [Google Scholar] [CrossRef]
- Camin, F.; Bontempo, L.; Perini, M.; Piasentier, E. Stable isotope ratio analysis for assessing the authenticity of food of animal origin. Compr. Rev. Food Sci. Food Saf. 2016, 15, 868–877. [Google Scholar] [CrossRef] [Green Version]
- Won, E.-J.; Kim, S.-H.; Go, Y.-S.; Kumar, K.S.; Kim, M.-S.; Yoon, S.-H.; Bayon, G.; Kim, J.-H.; Shin, K.-H. A multi-elements isotope approach to assess the geographic provenance of manila clams (Ruditapes philippinarum) via recombining appropriate elements. Foods 2021, 10, 646. [Google Scholar] [CrossRef]
- Kim, H.; Kumar, K.S.; Shin, K.-H. Applicability of stable C and N isotope analysis in inferring the geographical origin and authentication of commercial fish (Mackerel, Yellow Croaker and Pollock). Food Chem. 2015, 172, 523–527. [Google Scholar] [CrossRef]
- Watanabe, S.; Kodama, M.; Fukuda, M. Nitrogen stable isotope ratio in the manila clam, Ruditapes philippinarum, reflects eutrophication levels in tidal flats. Mar. Pollut. Bull. 2009, 58, 1447–1453. [Google Scholar] [CrossRef]
- Dang, C.; Sauriau, P.-G.; Savoye, N.; Caill-Milly, N.; Martinez, P. Determination of diet in Manila clams by spatial analysis of stable isotopes. Mar. Ecol. Prog. Ser. 2009, 387, 167–177. [Google Scholar] [CrossRef] [Green Version]
- Rubenstein, D.R.; Hobson, K.A. From birds to butterflies: Animal movement patterns and stable isotope. Trends Ecol. Evol. 2004, 19, 256–263. [Google Scholar] [CrossRef]
- Thomas, F.; Jamin, E.; Wietzerbin, K.; Guérin, R.; Lees, M.; Morvan, E.; Billault, I.; Derrien, S.; Rojas, J.M.M.; Serra, F.; et al. Determination of origin of Atlantic salmon (Salmo salar): The use of multiprobe and multielement isotopic analyses in combination with fatty acid composition to assess wild or farmed origin. J. Agric. Food Chem. 2008, 56, 989–997. [Google Scholar] [CrossRef]
- Ricardo, F.; Pimentel, T.; Moreira, A.S.; Rey, F.; Coimbra, M.A.; Domingues, M.R.; Domingues, P.; Leal, M.C.; Calado, R. Potential use of fatty acid profiles of the adductor muscle of cockles (Cerastoderma edule) for traceability of collection site. Sci. Rep. 2015, 5, 11125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Leeuwen, K.A.; Camin, F.; Jerónimo, E.; Vasta, V.; Prenzler, P.D.; Ryan, D.; Bessa, R.J. Dietary effects on stable carbon isotope composition of fatty acids in polar and neutral fractions of intramuscular fat of lambs. J. Agric. Food Chem. 2017, 65, 9404–9411. [Google Scholar] [CrossRef] [PubMed]
- Budge, S.M.; Iverson, S.J.; Koopman, H.N. Studying trophic ecology in marine ecosystems using fatty acids: A primer on analysis and interpretation. Mar. Mamm. Sci. 2006, 22, 759–801. [Google Scholar] [CrossRef]
- Kelly, J.R.; Scheibling, R.E. Fatty acids as dietary tracers in benthic food webs. Mar. Ecol. Prog. Ser. 2012, 446, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Nerot, C.; Meziane, T.; Schaal, G.; Grall, J.; Lorrain, A.; Paulet, Y.M.; Kraffe, E. Spatial changes in fatty acids signatures of the great scallop Pecten maximus across the Bay of Biscay continental shelf. Cont. Shelf Res. 2015, 109, 1–9. [Google Scholar] [CrossRef]
- Volkman, J.K.; Barrett, S.M.; Blackburn, S.I.; Mansour, M.P.; Sikes, E.L.; Gelin, F. Microalgal biomarkers: A review of recent research developments. Org. Geochem. 1998, 29, 1163–1179. [Google Scholar] [CrossRef]
- Surh, J.; Lee, H.-J.; Kwon, H. Regional difference in fatty acid content of Korean shellfish. Food Sci. Biotechnol. 2009, 18, 367–373. [Google Scholar]
- Burian, A.; Nielsen, J.M.; Hansen, T.; Bermudez, R.; Winder, M. The potential of fatty acid isotopes to trace trophic transfer in aquatic food-webs. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190652. [Google Scholar] [CrossRef]
- Magdas, D.A.; Guyon, F.; Puscas, R.; Vigouroux, A.; Gaillard, L.; Dehelean, A.; Feher, I.; Cristea, G. Applications of emerging stable isotopes and elemental markers for geographical and varietal recognition of Romanian and French honeys. Food Chem. 2021, 334, 127599. [Google Scholar] [CrossRef]
- Schlacher, T.A.; Connolly, R.M. Effect of acid treatment on carbon and nitrogen stable isotope ratios in ecological samples: A review and synthesis. Method Ecol. Evol. 2014, 5, 541–550. [Google Scholar] [CrossRef] [Green Version]
- Nichols, P.D.; Guckert, J.B.; White, D.C. Determination of monounsaturated fatty acid double-bond position and geometry for microbial monocultures and complex consortia by capillary GC-MS of their dimethyl disulphide adducts. J. Microbiol. Methods. 1986, 5, 49–55. [Google Scholar] [CrossRef]
- Lee, D.H.; Kim, J.H.; Lee, Y.M.; Jin, Y.K.; Paull, C.; Kim, D.; Shin, K.H. Chemosynthetic bacterial signatures in Frenulata tubeworm Oligobrachia sp. in an active mud volcano of the Canadian Beaufort Sea. Mar. Ecol. Prog. Ser. 2019, 628, 95–104. [Google Scholar] [CrossRef]
- Bayne, B.L. Phenotypic flexibility and physiological tradeoffs in the feeding and growth of marine bivalve molluscs. Integr. Comp. Biol. 2004, 44, 425–432. [Google Scholar] [CrossRef]
- Suh, Y.J.; Shin, K.H. Size-related and seasonal diet of the manila clam (Ruditapes philippinarum), as determined using dual stable isotopes. Estuar. Coast. Shelf Sci. 2013, 135, 94–105. [Google Scholar] [CrossRef]
- Komorita, T.; Kajihara, R.; Tsutsumi, H.; Shibanuma, S.; Yamada, T. Food sources for Ruditapes philippinarum in a coastal lagoon determined by mass balance and stable isotope approaches. PLoS ONE 2014, 9, e86732. [Google Scholar] [CrossRef]
- Kang, C.K.; Kim, J.B.; Lee, K.S.; Kim, J.B.; Lee, P.Y.; Hong, J.S. Trophic importance of benthic microalgae to macrozoobenthos in coastal bay systems in Korea: Dual stable C and N isotope analyses. Mar. Ecol. Prog. Ser. 2003, 259, 79–92. [Google Scholar] [CrossRef]
- Kang, C.K.; Choy, E.J.; Song, H.S.; Park, H.J.; Soe, I.S.; Jo, Q.T.; Lee, K.S. Isotopic determination of food sources of benthic invertebrates in two different macroalgal habitats in the Korean Coasts. J. Korean Soc. Oceanogr. 2007, 12, 380–389. (In Korean) [Google Scholar]
- Hobson, K.A. Stable-carbon and nitrogen isotope ratios of songbird feathers grown in two terrestrial biomes: Implications for evaluating trophic relationships and breeding origins. Condor 1999, 101, 799–805. [Google Scholar] [CrossRef] [Green Version]
- Elliott, K.H.; Braune, B.M.; Elliott, J.E. Byond bulk δ15N: Combining a suite of stable isotopic measures improves the resolution of the food webs mediating contaminant signals across space, time and communities. Environ. Int. 2021, 148, 106370. [Google Scholar] [CrossRef]
- Choi, B.; Shin, K.H. Determination of precise nitrogen stable isotopic baselines from heterotrophic organism in coastal ocean environments using compound specific isotope analysis of amino acids. Mar. Pollut. Bull. 2021, 171, 112777. [Google Scholar] [CrossRef]
- Fukumori, K.; Oi, M.; Doi, H.; Takahashi, D.; Okuda, N.; Miller, T.W.; Kuwae, M.; Miyasaka, H.; Genkai-kato, M.; Koizumi, Y.; et al. Bivalve tissue as a carbon and nitrogen isotope baseline indicator in coastal ecosystem. Estuar. Coastal Shelf Sci. 2008, 79, 45–50. [Google Scholar] [CrossRef]
- McClelland, J.W.; Valiela, I. Nitrogen-stable isotope signatures in estuarine food webs: A record of increasing urbanization in coastal watersheds. Limnol. Oveanogr. 1996, 42, 930–937. [Google Scholar] [CrossRef]
- Kim, T.-W.; Lee, K.; Lee, C.-K.; Jeong, H.-D.; Suh, Y.-S.; Lim, W.-A.; Kim, K.Y.; Jeong, H.-J. Interannual nutrient dynamics in Korean coastal waters. Harmful Algae 2013, 30, S15–S27. [Google Scholar] [CrossRef]
- Schmidt, O.; Quilter, J.M.; Bahar, B.; Moloney, A.P.; Scrimgeour, C.M.; Begley, I.S.; Monahan, F.J. Inferring the origin and dietary history of beef from C, N and S stable isotope ratio analysis. Food Chem. 2005, 91, 545–549. [Google Scholar] [CrossRef]
- Ocvirk, M.; Ogrinc, N.; Košir, I.J. Determination of the geographical and botanical origin of Hops (Humulus lupulus L.) using stable isotopes of C, N, and S. J. Agric. Food Chem. 2018, 66, 2021–2026. [Google Scholar] [CrossRef]
- Ricardo, F.; Maciel, E.; Domingues, M.R.; Calado, R. Spatio-temporal variability in the fatty acid profile of the adductor muscle of the common cockle Cerastoderma edule and its relevance for tracing geographic origin. Food Cont. 2017, 81, 173–180. [Google Scholar] [CrossRef]
- Andrea, C.; Alfaro, A.C.; Thomas, F.; Sergent, L.; Duxbury, M. Identification of trophic interactions within an estuarine food web (northern New Zealand) using fatty acid biomarkers and stable isotopes. Estuar. Coastal Shelf Sci. 2006, 70, 271–286. [Google Scholar]
- Gonçalves, A.M.M.; Marques, J.C.; Gonçalves, F. Fatty Acids’ Profiles of Aquatic Organisms: Revealing the Impacts of Environmental and Anthropogenic Stressors; Catala, A., Ed.; Intech: London, UK, 2016. [Google Scholar]
- Surh, J.; Kwon, H. Fatty acid contents and composition of various Korean shellfish. Food Sci. Biotechnol. 2003, 12, 83–87. [Google Scholar]
- Bergé, J.P.; Barnathan, G. Fatty acid from lipids of marine organisms: Molecular biodiversity, roles as biomarkers, biologically active compounds, and economical aspects. In Marine Biotechnology I; Springer: Berlin/Heidelberg, Germany, 2005; pp. 49–125. [Google Scholar]
- Parrish, C.C.; Deibel, D.; Thompson, R.J. Effect of sinking spring phytoplankton blooms on lipid content and composition in suprabenthic and benthic invertebrates in a cold ocean coastal environment. Mar. Ecol. Prog. Ser. 2009, 391, 33–51. [Google Scholar] [CrossRef]
- MOF. Coastal Ecosystem Survey Annual Report. Ministry of Oceans and Fishery. 2017. Available online: https://www.meis.go.kr/mes/data/1/board.do (accessed on 27 June 2022).
- Park, K. Variation of nitric oxide concentrations in response to shaking stress in the Manila clam Ruditapes philippinarum. Korean J. Malacol. 2013, 29, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Busetto, M.L.; Moretti, V.M.; Moreno-Rojas, J.M.; Caprino, F.; Giani, I.; Malandra, R.; Bellagamba, F.; Guillou, C. Authentication of farmed and wild turbot (Psetta maxima) by fatty acid and isotopic analyses combined with chemometrics. J. Agric. Food Chem. 2008, 56, 2742–2750. [Google Scholar] [CrossRef]
- Cossignani, L.; Pollini, L.; Blasi, F. Invited review: Authentication of milk by direct and indirect analysis of triacylglycerol molecular species. J. Dairy Sci. 2019, 102, 5871–5882. [Google Scholar] [CrossRef]
- González-Domínguez, R.; Sayago, A.; Fernández-Recamales, Á. Fatty acid profiling for the authentication of Iberian hams according to the feeding regime. Foods 2020, 9, 149. [Google Scholar] [CrossRef] [Green Version]
- Peycheva, K.; Panayotova, V.; Stancheva, R.; Makedonski, L.; Merdzhanova, A.; Cicero, N.; Parrino, V.; Fazio, F. Trace elements and omega-3 fatty acids of wild and farmed Mussels (Mytilus galloprovincialis) consumed in Bulgaria: Human health risks. Int. J. Environ. Res. Public Health 2021, 18, 10023. [Google Scholar] [CrossRef]
- Twining, C.W.; Taipale, S.J.; Ruess, L.; Bec, A.; Martin-Creuzburg, D.; Kainz, M.J. Stable isotopes of fatty acids: Current and future perspectives for advancing trophic ecology. Phil. Trans. R. Soc. 2020, 375, 20190641. [Google Scholar] [CrossRef]
- Howland, M.R.; Corr, L.T.; Young, S.M.M.; Jones, V.; Jim, S.; Van der Merwe, N.J.; Mitchell, A.D.; Evershed, R.P. Expression of the dietary isotope signal in the compound-specific d13C values of pig bone lipids and amino acids. Int. J. Osteoarchaeol. 2003, 13, 54–65. [Google Scholar] [CrossRef]
- Yun, H.Y.; Larsen, T.; Choi, B.; Won, E.J.; Shin, K.H. Amino acid nitrogen and carbon isotope data: Potential and implications for ecological studies. Ecol. Evol. 2022, 12, e8929. [Google Scholar] [CrossRef]
- Chung, I.M.; Kim, Y.J.; Moon, H.S.; Kwon, C.; Chi, H.Y.; Kim, S.H. Regional Characterization Study of Fatty Acids and Tocopherol in Organic Milk as a Tool for Potential Geographical Identification. Foods 2020, 9, 1743. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, S.; Jin, X. Fatty acid stable carbon isotope ratios combined with oxidation kinetics for characterization and authentication of walnut oils. J. Agr. Food Chem. 2021, 23, 6701–6709. [Google Scholar] [CrossRef]
- Zhang, X.; Han, D.; Chen, X.; Zhao, X.; Cheng, J.; Liu, Y. Combined use of fatty acid profile and fatty acid δ13C fingerprinting for origin traceability of scallops (Patinopecten yessoensis, Chlamys farreri, and Argopecten irradians). Food Chem. 2019, 298, 124966. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, J.; Li, D.J.; Wei, H.; Lu, R.X. Isotope variability of particulate organic matter at the PN section in the East China Sea. Biogeochemistry 2003, 65, 31–49. [Google Scholar] [CrossRef]
- Yoo, M.H.; Park, K.W.; Oh, H.J.; Koo, J.H.; Kwon, J.N.; Youn, S.H. The seasonal and regional distribution of phytoplankton communities in the fisheries resources protection area of Korea in 2016. J. Korean Soc. Mar. Environ. Saf. 2008, 24, 288–293. [Google Scholar] [CrossRef]
- Liu, M.; Li, C.; Sun, S. Seasonal variation in fatty acid composition of seston and the copepod Calanus sinicus (Brodsky, 1962) in Jiaozhou Bay and its trophic implications. Chin. J. Oceanol. Limnol. 2011, 29, 1164–1173. [Google Scholar] [CrossRef]






| Region | ID | δ13C (‰) | δ15N (‰) | Remarks (Sites, Sample N) | Sampling Date |
|---|---|---|---|---|---|
| Chungcheong | CC-1 | −18.0 ± 0.2 | 8.9 ± 0.2 | Dangjin (37.035906° N, 126.573153° E, n = 5) | 21 July 2016 |
| (n = 25) | CC-2 | −18.3 ± 0.3 | 6.8 ± 0.3 | Sogeunri (36.816236° N, 126.144142° E, n = 5) | 21 June 2016 |
| CC-3 | −18.4 ± 0.1 | 6.4 ± 0.4 | Uihang (36.887164° N, 126.372181° E, n = 5) | 21 June 2016 | |
| CC-4 | −17.6 ± 0.4 | 9.1 ± 0.2 | Hongseong (36.519644° N, 126.484239° E, n = 5) | 22 June 2016 | |
| CC-5 | −17.5 ± 0.3 | 8.6 ± 0.4 | Boryeong (36.232231° N, 126,509742° E, n = 5) | 23 June 2016 | |
| Average | −18.0 ± 0.4 a | 7.9 ± 1.2 a | |||
| Jeonbuk | JB-1 | −17.1 ± 0.1 | 9.6 ± 0.4 | Saemangeum (35.836264° N, 126.544094° E, n = 10) | 24 June 2016 |
| (n = 25) | JB-2 | −16.6 ± 0.3 | 9.1 ± 0.3 | Wedo (35.576103° N, 126.254097° E, n = 5) | 21 June 2016 |
| JB-3 | −17.4 ± 0.1 | 10.0 ± 0.3 | Gochang (35.549975° N, 126.559472° E, n = 5) | 24 June 2016 | |
| JB-4 | −16.4 ± 0.4 | 8.7 ± 0.4 | Shinan (35.103347° N, 126.1363342° E, n = 5) | 20 July 2016 | |
| Average | −16.9 ± 0.4 b | 9.4 ± 0.6 c | |||
| Jeonnam | JN-1 | −16.5 ± 0.3 | 7.7 ± 0.1 | Wando (34.278747° N, 126.729958° E, n = 10) | 14 June 2016 |
| (n = 26) | JN-2 | −17.1 ± 0.1 | 10.0 ± 0.2 | Hwayang (34.688503° N, 127.600969° E, n = 3) | 24 July 2015 |
| JN-3 | −16.9 ± 0.2 | 9.0 ± 0.1 | Dolsan (34.669314° N, 127.766706° E, n = 3) | 29 July 2015 | |
| JN-4 | −16.9 ± 0.2 | 9.1 ± 0.1 | Yeosu (34.614800° N, 237.676294° E, n = 10) | 26 May 2016 | |
| Average | −16.8 ± 0.3 b | 8.7 ± 0.8 b | |||
| Gyeongnam | GN-1 | −16.8 ± 0.3 | 8.8 ± 0.2 | Tongyeong (34.733336° N, 128.386897° E, n = 8) | 14 July 2016 |
| (n = 26) | GN-2 | −15.7 ± 0.1 | 10.6 ± 0.2 | Geoje (34.732281° N, 128.691311° E, n = 10) | 20 May 2016 |
| GN-3 | −16.3 ± 0.1 | 9.8 ± 0.2 | Masan (35.096711° N, 128.672411° E, n = 3) | 24 July 2015 | |
| GN-4 | −15.8 ± 0.2 | 11.0 ± 0.1 | Udo (35.087564° N, 128.720750° E, n = 5) | 15 July 2016 | |
| Average | −16.1 ± 0.6 c | 10.0 ± 0.9 c |
| Region | Number of Samples Classified by LDA | Prediction | |||
|---|---|---|---|---|---|
| CC | GN | JB | JN | (%) | |
| Chungcheong (CC) | 21 | 0 | 3 | 1 | 84.0 |
| Gyeongnam (GN) | 0 | 19 | 2 | 5 | 73.1 |
| Jeonbuk (JB) | 0 | 2 | 15 | 8 | 60.0 |
| Jeonnam (JN) | 0 | 0 | 12 | 14 | 53.8 |
| Region | Number of Samples Classified by LDA | Prediction | |||
|---|---|---|---|---|---|
| CC | GN | JB | JN | (%) | |
| Chungcheong (CC) | 15 | 0 | 0 | 0 | 100 |
| Gyeongnam (GN) | 0 | 12 | 0 | 0 | 100 |
| Jeonbuk (JB) | 0 | 0 | 12 | 0 | 100 |
| Jeonnam (JN) | 0 | 0 | 0 | 12 | 100 |
| Regions | ID | δ13C16:0 | δ13C18:0 | δ13C18:1n-7 | δ13C18:1n-9 | δ13C20:1n-7 | δ13C20:1n-9 |
|---|---|---|---|---|---|---|---|
| Chungcheong | CC-1 | −23.1 ± 0.4 | −24.6 ± 0.4 | −20.9 ± 0.8 | −15.2 ± 0.7 | −22.9 ± 0.5 | −21.0 ± 0.7 |
| (n = 15) | CC-2 | −24.4 ± 0.4 | −25.3 ± 0.4 | −22.5 ± 0.4 | −15.6 ± 0.5 | −25.0 ± 0.3 | −22.4 ± 0.3 |
| CC-3 | −24.4 ± 0.4 | −25.3 ± 0.4 | −22.2 ± 0.3 | −15.5 ± 0.4 | −24.6 ± 0.3 | −21.3 ± 0.5 | |
| CC-4 | −25.0 ± 0.5 | −25.3 ± 0.3 | −21.1 ± 0.7 | −18.5 ± 0.5 | −20.7 ± 0.5 | −19.9 ± 0.3 | |
| CC-5 | −24.9 ± 0.4 | −25.0 ± 0.3 | −20.2 ± 0.4 | −17.9 ± 0.4 | −20.6 ± 0.6 | −20.4 ± 0.5 | |
| Avg. | −24.3 ± 0.8 a | −25.1 ± 0.4 a | −21.4 ± 1.0 b | −16.5 ± 1.5 b | −22.8 ± 2.0 b | −21.0 ± 1.0 bc | |
| Jeonbuk | JB-1 | −20.4 ± 0.5 | −24.5 ± 0.4 | −20.6 ± 0.5 | −15.6 ± 0.5 | −21.0 ± 0.4 | −20.9 ± 0.4 |
| (n = 12) | JB-2 | −21.9 ± 0.4 | −21.4 ± 0.8 | −21.4 ± 0.5 | −15.0 ± 0.7 | −21.1 ± 0.9 | −21.6 ± 0.5 |
| JB-3 | −26.8 ± 0.4 | −26.1 ± 0.8 | −24.5 ± 0.7 | −19.6 ± 0.5 | −24.8 ± 0.6 | −21.7 ± 0.6 | |
| JB-4 | −23.5 ± 0.4 | −21.2 ± 0.3 | −21.2 ± 0.4 | −17.5 ± 0.5 | −21.5 ± 0.4 | −23.6 ± 0.5 | |
| Avg. | −23.1 ± 2.5 ab | −23.3 ± 2.2 b | −21.9 ± 1.7 b | −16.9 ± 1.9 ab | −22.1 ± 1.7 bc | −21.9 ± 1.1 ab | |
| Jeonnam | JN-1 | −22.2 ± 0.4 | −19.8 ± 0.3 | −20.7 ± 0.5 | −16.8 ± 0.5 | −20.6 ± 0.5 | −21.3 ± 0.5 |
| (n = 12) | JN-2 | −23.2 ± 0.3 | −19.8 ± 0.4 | −22.0 ± 0.5 | −18.0 ± 0.4 | −21.5 ± 0.5 | −23.1 ± 0.5 |
| JN-3 | −23.4 ± 0.4 | −21.2 ± 0.6 | −22.6 ± 0.5 | −18.0 ± 0.4 | −20.9 ± 0.6 | −23.3 ± 0.4 | |
| JN-4 | −23.5 ± 0.3 | −20.6 ± 0.5 | −22.1 ± 0.6 | −18.0 ± 0.4 | −21.8 ± 0.4 | −23.4 ± 0.5 | |
| Avg. | −23.1 ± 0.6 ab | −20.3 ± 0.7 c | −21.9 ± 0.9 b | −17.7 ± 0.7 ab | −21.2 ± 0.7 c | −22.8 ± 1.0 a | |
| Gyeongnam | GN-1 | −24.5 ± 0.6 | −24.2 ± 0.6 | −27.1 ± 0.7 | −19.5 ± 0.6 | −26.1 ± 0.5 | −23.6 ± 0.6 |
| (n = 12) | GN-2 | −21.6 ± 0.5 | −22.1 ± 0.3 | −25.6 ± 0.6 | −17.8 ± 0.6 | −24.6 ± 0.5 | −18.0 ± 0.6 |
| GN-3 | −21.9 ± 0.3 | −22.2 ± 0.4 | −25.5 ± 0.6 | −17.8 ± 0.3 | −24.8 ± 0.3 | −18.0 ± 0.5 | |
| GN-4 | −22.0 ± 0.5 | −22.2 ± 0.7 | −25.7 ± 0.5 | −17.8 ± 0.4 | −24.8 ± 0.5 | −18.1 ± 0.3 | |
| Avg. | −22.5 ± 1.3 b | −22.7 ± 1.0 b | −26.0 ± 0.9 a | −18.2 ± 0.9 a | −25.1 ± 0.7 a | −19.4 ± 2.6 c |
| Region | Number of Samples Classified by LDA (Cross-Validation) | Prediction | |||
|---|---|---|---|---|---|
| CC | GN | JB | JN | (%) | |
| Chungcheong (CC) | 14 | 0 | 1 | 0 | 93.3 |
| Gyeongnam (GN) | 0 | 12 | 0 | 0 | 100 |
| Jeonbuk (JB) | 1 | 0 | 7 | 4 | 58.3 |
| Jeonnam (JN) | 0 | 0 | 0 | 12 | 100 |
| Country | ID | δ13C (‰) | δ15N (‰) | Sampling Date * |
|---|---|---|---|---|
| China (Dalian) | CHN | −17.58 ± 0.39 b | 9.67 ± 0.72 a | 1 September 2015 |
| DPR Korea | NK | −17.69 ± 0.38 b | 8.95 ± 0.24 a | 1 September 2015 |
| Korea | KOR | −16.94 ± 0.79 a | 9.01 ± 1.18 a | - |
| Country | Number of Samples Correctly Classified | ||
|---|---|---|---|
| Step 1. δ13C, δ15N | Step 2-I. FA Profiles | Step 2-II. δ13C-FA | |
| China (CHN) | 15 (4) | 3 (3) | 3 (3) |
| Korea (KOR) | 102 (98) | 51 (51) | 51 (51) |
| DPR Korea (NK) | 16 (1) | 3 (2) | 3 (3) |
| Total (Cross-Val)% | 77.44% | 98.24% | 100% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Go, Y.-S.; Won, E.-J.; Kim, S.-H.; Lee, D.-H.; Kang, J.-H.; Shin, K.-H. Stepwise Approach for Tracing the Geographical Origins of the Manila Clam Ruditapes philippinarum Using Dual-Element Isotopes and Carbon Isotopes of Fatty Acids. Foods 2022, 11, 1965. https://doi.org/10.3390/foods11131965
Go Y-S, Won E-J, Kim S-H, Lee D-H, Kang J-H, Shin K-H. Stepwise Approach for Tracing the Geographical Origins of the Manila Clam Ruditapes philippinarum Using Dual-Element Isotopes and Carbon Isotopes of Fatty Acids. Foods. 2022; 11(13):1965. https://doi.org/10.3390/foods11131965
Chicago/Turabian StyleGo, Young-Shin, Eun-Ji Won, Seung-Hee Kim, Dong-Hun Lee, Jung-Ha Kang, and Kyung-Hoon Shin. 2022. "Stepwise Approach for Tracing the Geographical Origins of the Manila Clam Ruditapes philippinarum Using Dual-Element Isotopes and Carbon Isotopes of Fatty Acids" Foods 11, no. 13: 1965. https://doi.org/10.3390/foods11131965
APA StyleGo, Y. -S., Won, E. -J., Kim, S. -H., Lee, D. -H., Kang, J. -H., & Shin, K. -H. (2022). Stepwise Approach for Tracing the Geographical Origins of the Manila Clam Ruditapes philippinarum Using Dual-Element Isotopes and Carbon Isotopes of Fatty Acids. Foods, 11(13), 1965. https://doi.org/10.3390/foods11131965